|
Size: 2731
Comment:
|
Size: 3818
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| ## page was renamed from CancerStemCellProject/ChangjiangXu/BiostatService ##master-page:HomepageTemplate #format wiki |
|
| Line 8: | Line 5: |
| The Biostatistics Service is freely available to all Cancer Stem Cell program members. You can get help with the design and analysis of genomics experiments, for example, sample size, biological replicates, or pooled samples. | The Biostatistics Service is freely available to all Cancer Stem Cell program members. |
| Line 11: | Line 8: |
| * We provide help with design issues of genomics experiments (e.g. gene expression microarrays, RNA-Seq), such as how to structure your samples to avoid confounding factors that will make your analysis more difficult and reduce power of your analyses. To help you with statistical planning of your experiment, we need clear and concise information about your plans. A document related to the type of information that we need to understand your experiment is provided in [[CSCBiostatService/Consulting| CONSULTING]]. | * We provide help with design issues of genomics experiments (e.g. gene expression microarrays, RNA-Seq), such as how to structure your samples to avoid confounding factors that will make your analysis more difficult and reduce power of your analyses. To help you with statistical planning of your experiment, we need clear and concise information about your plans. A document related to the type of information that we need to understand your experiment is provided in [[CSCBiostatService/Consulting | CONSULTING]]. |
| Line 13: | Line 10: |
== Statistical Analysis == * We provide statistical analysis, e.g., normalization, batch correction, or model fitting. The statistical analysis is available for any problem related to the cancer stem cell network: * Statistical analysis of different experiment data (or other omics data): * [[CSCBiostatService/AffymetrixDataAnalysis | Affymetrix microarray data]] * [[CSCBiostatService/IlluminaChipDataAnalysis | Illumina beadchip data]] * RNA-Seq data * Flow cytometry data * Multiple testing adjustments and calculation of false discovery rates. * Meta-analysis of multiple studies * Survival analysis, multivariate analysis, etc, and other problems as needed == Consulting and Training == * If you are doing the analyses yourself but you need advice, please feel free to contact us for a consulting meeting at any stage of your analysis. == Expectations and Data Requirements == |
|
| Line 17: | Line 31: |
| == Statistical Analysis == * We also provide statistical analysis support, for example, normalization, batch correction, or model fitting * Affymetrix microarray data * Illumina beadchip data * RNA-Seq data |
|
| Line 24: | Line 33: |
| * If you need help with data interpretation (Pathway and Network Analysis), you can contact Dr. Veronique Voisin (Ph.D, bioinformatician) at veronique.voisin@gmail.com. | * Once data from your genomics experiment have been statistically analyzed, the end result is a list of genes that can contain only a few genes or more frequently hundreds or thousands. These genes are likely to be related to the condition or physiological perturbation that you are studying. The next step is to functionally interpret these data by integrating prior knowledge about these genes in an automatic manner or by integrating as a network several layers of information or data that you can have. We have developed a pathway and network analysis service (http://www.baderlab.org/CSCPathwayAnalysisService) which goal is to help with data interpretation. Please, do not hesitate to contact Dr. Veronique Voisin (veronique.voisin@gmail.com) for more information about this service or visit the website. |
| Line 26: | Line 35: |
| == Useful Software Tools == | == Useful Software Tools == |
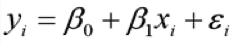
OICR Cancer Stem Cell Program - Biostatistics Service
The Biostatistics Service is freely available to all Cancer Stem Cell program members.
Experiments Design
We provide help with design issues of genomics experiments (e.g. gene expression microarrays, RNA-Seq), such as how to structure your samples to avoid confounding factors that will make your analysis more difficult and reduce power of your analyses. To help you with statistical planning of your experiment, we need clear and concise information about your plans. A document related to the type of information that we need to understand your experiment is provided in CONSULTING.
Statistical Analysis
- We provide statistical analysis, e.g., normalization, batch correction, or model fitting. The statistical analysis is available for any problem related to the cancer stem cell network:
- Statistical analysis of different experiment data (or other omics data):
- RNA-Seq data
- Flow cytometry data
- Multiple testing adjustments and calculation of false discovery rates.
- Meta-analysis of multiple studies
- Survival analysis, multivariate analysis, etc, and other problems as needed
- Statistical analysis of different experiment data (or other omics data):
Consulting and Training
- If you are doing the analyses yourself but you need advice, please feel free to contact us for a consulting meeting at any stage of your analysis.
Expectations and Data Requirements
If you have data from genomics (e.g. gene expression) experiment, we need certain information to understand and do efficient analysis of your data which are documented in DATA REQUIREMENTS.
For information regarding what you should expect from the service, please read EXPECTATIONS.
Data Interpretation
Once data from your genomics experiment have been statistically analyzed, the end result is a list of genes that can contain only a few genes or more frequently hundreds or thousands. These genes are likely to be related to the condition or physiological perturbation that you are studying. The next step is to functionally interpret these data by integrating prior knowledge about these genes in an automatic manner or by integrating as a network several layers of information or data that you can have. We have developed a pathway and network analysis service (http://www.baderlab.org/CSCPathwayAnalysisService) which goal is to help with data interpretation. Please, do not hesitate to contact Dr. Veronique Voisin (veronique.voisin@gmail.com) for more information about this service or visit the website.
Useful Software Tools
VLOOKUP: After the analysis of a gene expression experiment, often you have a few Excel spreadsheets and want to find out which genes are common between the two spread sheets. With large data sets, this can be a daunting task. Efficient use of Excel function VLOOKUP can help you find commonalities between the two spreadsheets. The VLOOKUP function can also be used to pull information from one spreadsheet on the basis of some reference values from the other spreadsheet & concatenate the set of information with the 2nd spreadsheet. For detailed instructions, please download the vlookup_new.xlsx file instead of viewing it in web browser.
Contact Dr. Changjiang Xu (Biostatistician) changjiang.xu@utoronto.ca
