|
Size: 345
Comment:
|
Size: 2441
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| {{{ ##master-page:HomepageTemplate #format wiki == Changjiang Xu == Email: changjiang.xu@utoronto.ca |
#acl CscGroup:read,write,revert |
| Line 7: | Line 3: |
| * [[CancerStemCellProject/ChangjiangXu/ProjectCSC|Cancer Stem Cell]] | {{attachment:logobiostat.png|OICR_CSC Pathway and Network Analysis Logo Map Logo|align="right"}} <<BR>> = OICR CSC Statistical Analysis Service = |
| Line 9: | Line 6: |
| * [[CancerStemCellProject/ChangjiangXu/BiostatService|Biostatistics Service]] | == OICR Cancer Stem Cell program == |
| Line 11: | Line 8: |
| Changjiang Xu, Gary Bader ------ * [[ChangjiangXu/Intranet/Weekly | Weekly Updates]] * OICR CSC Projects (PI's name and/or project title listed below) * [[CancerStemCellProject/ChangjiangXu/LA02 | Laurie Ailles, Crosstalk between CAFs and ovarian cancer cells (LA02_CAF_ovarian_Laurie) ]] * [[CancerStemCellProject/ChangjiangXu/PD03 | Peter Dirks, Role of dopamine D4 receptor in glioblastoma stem cells]] * [[CancerStemCellProject/ChangjiangXu/CG04 | Cyndi Guidos]] * [[CancerStemCellProject/ChangjiangXu/oPOSSUM | oPOSSUM]] * HALT02/JAK2 * [[CancerStemCellProject/ChangjiangXu/HALT02_SRC | Differential expression analysis of SRC high vs low (negative) sub-populations]] * [[CancerStemCellProject/ChangjiangXu/HALT02_JAK2 | Comparison of the pathways for XG-R/PF-R, XG-NR/PF-NR, and XG-R/PF-NR]] * [[CancerStemCellProject/ChangjiangXu/KH01 | Kristin Hope]] * [[CancerStemCellProject/ChangjiangXu/BP03 | Bret Pearson, Spatial Organization of the planarian transcriptome]] * [[CancerStemCellProject/ChangjiangXu/BP04 | Bret Pearson, Mechanisms driving asymmetric cell fate outcomes in planarian adult stem cell lineages]] * [[CancerStemCellProject/VeroniqueVoisin/BiostatisticProjects | Potential Biostatistics Projects from Bioinformatics Service]] * [[CSCBiostatService | Biostatistics Service]] * Software Tools * GSEA (see [[CSCBiostatService/GSEA | a summary of GSEA methods and software]]) * '''VLOOKUP''': After the analysis of a gene expression experiment, often you have a few Excel spreadsheets and want to find out which genes are common between the two spread sheets. With large data sets, this can be a daunting task. Efficient use of Excel function VLOOKUP can help you find commonalities between the two spreadsheets. The VLOOKUP function can also be used to pull information from one spreadsheet on the basis of some reference values from the other spreadsheet & concatenate the set of information with the 2nd spreadsheet. For detailed instructions, please download [[attachment:vlookup_new.xlsx| the vlookup_new.xlsx]] file instead of viewing it in web browser. * [[CSCPathwayAnalysisService/Tutorials | More]] |
|
| Line 13: | Line 35: |
| CategoryHomepage }}} Describe CancerStemCellProject/ChangjiangXu here. |
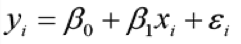
OICR CSC Statistical Analysis Service
OICR Cancer Stem Cell program
Changjiang Xu, Gary Bader
- OICR CSC Projects (PI's name and/or project title listed below)
Laurie Ailles, Crosstalk between CAFs and ovarian cancer cells (LA02_CAF_ovarian_Laurie)
Peter Dirks, Role of dopamine D4 receptor in glioblastoma stem cells
- HALT02/JAK2
Bret Pearson, Spatial Organization of the planarian transcriptome
Bret Pearson, Mechanisms driving asymmetric cell fate outcomes in planarian adult stem cell lineages
Potential Biostatistics Projects from Bioinformatics Service
- Software Tools
GSEA (see a summary of GSEA methods and software)
VLOOKUP: After the analysis of a gene expression experiment, often you have a few Excel spreadsheets and want to find out which genes are common between the two spread sheets. With large data sets, this can be a daunting task. Efficient use of Excel function VLOOKUP can help you find commonalities between the two spreadsheets. The VLOOKUP function can also be used to pull information from one spreadsheet on the basis of some reference values from the other spreadsheet & concatenate the set of information with the 2nd spreadsheet. For detailed instructions, please download the vlookup_new.xlsx file instead of viewing it in web browser.
