|
Size: 2302
Comment:
|
← Revision 20 as of 2009-11-11 20:43:09 ⇥
Size: 658
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| #acl All:read | |
| Line 3: | Line 4: |
| == GeneMania DataWarehouse (DW) Related Documentation == | == GeneMania Data Warehouse (DW) == . The following list represents different components of the GeneMania Data warehouse subsystem: |
| Line 5: | Line 7: |
| DW Resource Loading: Entrez: ---------------------------- |
* [[GeneMania/DWResources|Data Warehouse Resources]] |
| Line 8: | Line 9: |
| The Entrez database, by NCBI, is released in two main formats. | * [[GeneMania/DWBuild|Data Warehouse Build/Comments]] |
| Line 10: | Line 11: |
| 1) Flat file format: This is composed of 11 main flat files, in a tab delimited format. The files are: gene2accession gene2go gene2pubmed gene2refseq gene2sts gene2unigene gene_history gene_info mim2gene gene_refseq_uniprotkb_collab interactions |
* [[GeneMania/IDMapping|Identifier Mapping]] |
| Line 23: | Line 13: |
| Most of these files are good for cross referencing and matching identifiers between different databases within NCBI and elsewhere. For example, matching a gene ID to the appropriate RNA/protein sequence IDs (gene2accession), to the published journal references (gene2pubmed), or to the associated human genetic diseases (mim2gene). Some of these files, however, have more meat in them, like gene2go (matching genes with GO ontologies), gene_info, and interactions (lists interactions with BIND, BioGrid, EcoCyc, HPRD). | * [[GeneMania/IDValidation|Identifier Validation]] |
| Line 25: | Line 15: |
| The local Entrez mirror is currently based on this format. The table/column names were purposefully matched to the file/header names for ease of use (except in cases where this might cause technical hassle, like having dots or spaces in column names). Note that there are no null columns in these tables. A hyphen '-' (and sometimes a '?') is usually used by the source files, instead. | * [[GeneMania/DWArchitect|Data Warehouse Architecture]] |
| Line 27: | Line 17: |
| The advantage of this format is ease of use, and the fact that the files are inclusive of all species information available from NCBI. Local views for the subsets of interest can be created as well. | * [[GeneMania/Ontology|Ontology Support]] |
| Line 29: | Line 19: |
| 2) ASN.1 Binary format: This compressed format can be transformed to XML using a program associated with the Entrez release as well. The structure of the bulky XML produced is based on DTDs by NCBI, and not on an XML schema. It is mainly split on a per species (or class) basis, but there are all inclusive files as well. It can be compared to the gene_info table listed above. | === More DW Documents: === [[GeneMania/GeneManiaDataCollection|GeneMANIA Data Collection]] |
| Line 31: | Line 22: |
| 3) There are other released files as well, that are of less significance or represent different subsets for the data listed above, like releasing the interactions for HIV as a separate file, and so on. 4) For more information, visit the Entrez FTP site at: ftp://ftp.ncbi.nlm.nih.gov/gene/ |
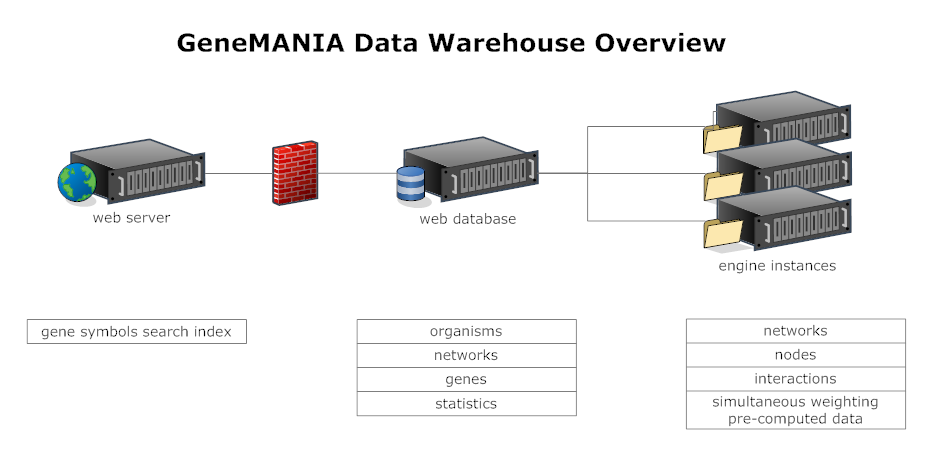
{{attachment:GeneManiaDataWarehouseOverview.png}} |
GeneMania Data Warehouse (DW)
The following list represents different components of the GeneMania Data warehouse subsystem:
* Data Warehouse Build/Comments
More DW Documents: