|
Size: 13171
Comment:
|
Size: 16228
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| #acl All:read | |
| Line 9: | Line 10: |
| * AutoAnnotate version 1.1 | |
| Line 15: | Line 17: |
| * Install AutoAnnotate | * Install AutoAnnotate (latest version is 1.1) |
| Line 149: | Line 151: |
| * Manage Annotation Sets... * Opens a Dialog where you can: * Change the order that Annotation Sets appear in the dropdown. * Delete multiple Annotation Sets at a time. |
|
| Line 151: | Line 157: |
| * Warning: Collapsing all clusters in a large network can be very slow. Consider using the "Create Summary Network" option instead. | |
| Line 153: | Line 160: |
| * Create Summary Network * Generates a new network where each node represents a collapsed cluster in the original network. * Very similar in functionality to "Collapse All", but runs significantly faster. |
|
| Line 158: | Line 168: |
| * Recalculates the labels for the selected clusters. Typically used when the WordCloud options or label algorithm changes. More on this later in this guide. | * Recalculates the labels for all of the clusters in the annotation set. Typically used when the WordCloud options or label algorithm changes. More on this later in this guide. (Note: to recalculate labels for a subset of clusters please select the clusters in the table and right-click to show the cluster context menu.) |
| Line 192: | Line 202: |
| * Warning: can be very slow when many clusters are selected. | |
| Line 194: | Line 205: |
| * Generate Summary Network * Generates a new network where each node represents a collapsed cluster that is selected in the original network. * Very similar in functionality to "Collapse", but runs significantly faster. |
|
| Line 211: | Line 225: |
| Clusters can be collapsed or expanded from the '''Annotation Set Menu''' or the '''Cluster Table Context Menu'''. | AutoAnnotate can summarize a network by collapsing some or all of the clusters automatically. Warning, as of Cytoscape 3.4 this function can be very slow, depending on the number of clusters. Consider using the "Generate Summary Network" option instead because it runs significantly faster. See the section below this one for more details. |
| Line 215: | Line 231: |
| * Before collapsing clusters make sure that node aggregation is enabled. If not enabled then collapsed clusters may not have a label. | * The Cytoscape "group nodes" feature is used to collapse and expand clusters. A group node is a compound node that contains other nodes. * When AutoAnnotate collapses a cluster it first creates a group node that contains all the nodes in the cluster and then the group node is collapsed. When the cluster is collapsed Cytoscape will create "meta-edges" between the group node any other nodes it is connected to. The collapsed group nodes and the meta-edges provide a summary of the network. * The annotation for a cluster is not shown when the cluster is collapsed. The label shown for a collapsed cluster is generated by Cytoscape and depends on the Group Preferences (explained below). * When a cluster is expanded the group node is expanded and then deleted. The annotations are show again. * It is not recommend to manually collapse or expand clusters using the Groups menu as this may confuse AutoAnnotate. * When switching between Annotation Sets all collapsed clusters are automatically expanded. * Clusters can be collapsed or expanded from the '''Annotation Set Menu''' or the '''Cluster Table Context Menu'''. * Before collapsing clusters make sure that node aggregation is enabled. This will tell Cytoscape to generate a label for the group node that aggregates the labels of the nodes in the cluster. |
| Line 221: | Line 245: |
| * When a cluster is collapsed a group node is created that contains the cluster and it is collapsed. The annotation for the cluster is not shown when the cluster is collapsed. * When a cluster is expanded the group node is expanded and then deleted. The annotation is show again. * It is not recommend to manually collapse or expand clusters using the Groups menu as this may confuse AutoAnnotate. * When switching between Annotation Sets all collapsed clusters are automatically expanded. |
== Summary Network == Using group nodes to collapse clusters can be very slow when there are many clusters. As an alternative a new option called '''Generate Summary Network''' has been added in AutoAnnotate 1.1. A new network is generated where each node represents a collapsed cluster in the original network, and each edge represents a meta-edge between clusters. The resulting network is very similar to the results you get from collapsing the clusters. The main differences are: * Nodes that are not contained in any cluster are ignored. * There is no relationship maintained back to the original network. If the clusters in the original network change the summary network will not be updated. Create a new summary network at any time to reflect the current state of the clusters. * A new visual style for the summary network is created based on the original visual style. * A summary network can be created from the '''Annotation Set Menu''' or the '''Cluster Table Context Menu'''. * Uses the same attribute aggregation settings as collapsed group nodes. Available from '''Edit > Preferences > Group Preferences''' then expand the '''Default Aggregation Settings''' section. |
| Line 239: | Line 271: |
| * When you restore a session it defaults back to (none) for each Network View. Simply select the annotation set again to redraw the annotations. |

AutoAnnotate: User Guide
Contents
-
AutoAnnotate: User Guide
- Getting Started (Installing)
- Creating an Annotation Set
- ClusterMaker2 Algorithms
- Panels
- Changing Display Options
- Managing Annotation Sets
- Managing Clusters in the AutoAnnotate Panel
- Managing Clusters in the Network View
- Collapsing and Expanding Clusters
- Summary Network
- Working with WordCloud
- Saving and Restoring a Session with Annotation Sets
- Additional Options
- Filing Bug Reports and Requesting Enhancements
Getting Started (Installing)
Requirements
- Cytoscape minimum version 3.3
AutoAnnotate version 1.1
WordCloud minimum version 3.1.0
- clusterMaker2 minimum version 0.9.5
Installation
- Install Cytoscape
If you don’t have Cytoscape please download and install the latest release from http://www.cytoscape.org/download.php.
Install AutoAnnotate (latest version is 1.1)
- Open Cytoscape
In the main menu select Apps > App Manager
In the App Manager select AutoAnnotate in the list of All Apps and click the Install button.
The WordCloud app will also be installed automatically along with AutoAnnotate
- Install clusterMaker2 (optional, but highly recommended)
In the App Manager select clusterMaker2 in the list of All Apps and click the Install button.
Alternatively AutoAnnotate and clusterMaker2 can be installed from the App Store website.
AutoAnnotate on the App Store: http://apps.cytoscape.org/apps/autoannotate
clusterMaker2 on the App Store: http://apps.cytoscape.org/apps/clustermaker2
Creating an Annotation Set
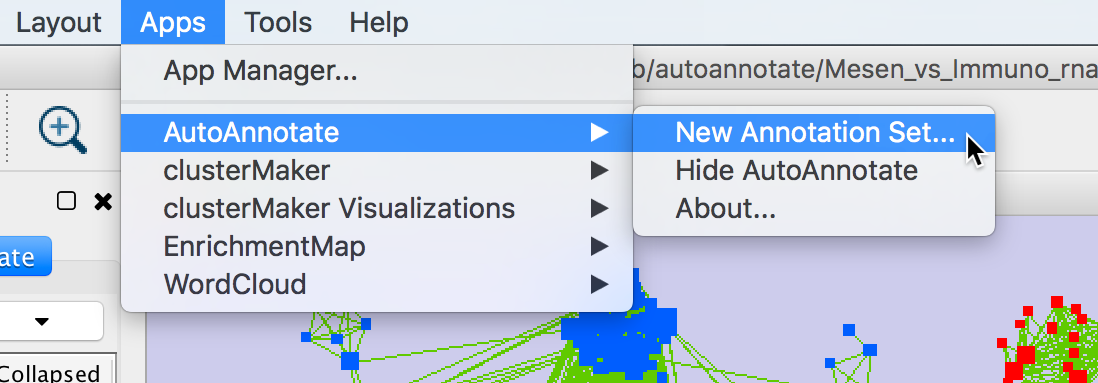
- Start by selecting a network view.
In the main menu select Apps > AutoAnnotate > New Annotation Set…
The Create Annotation Set dialog will open.
Create Annotation Set Dialog
- This dialog is used to select the options for creating an Annotation Set.
Note: AutoAnnotate can detect if the network was created by EnrichmentMap, and if so sensible defaults for EnrichmentMap networks will be pre-selected.
- Note: You may manually edit the contents and labels of the clusters after the Annotation Set is created, and you may create as many Annotation Sets as you like, so its not critical to get everything perfect at this step.
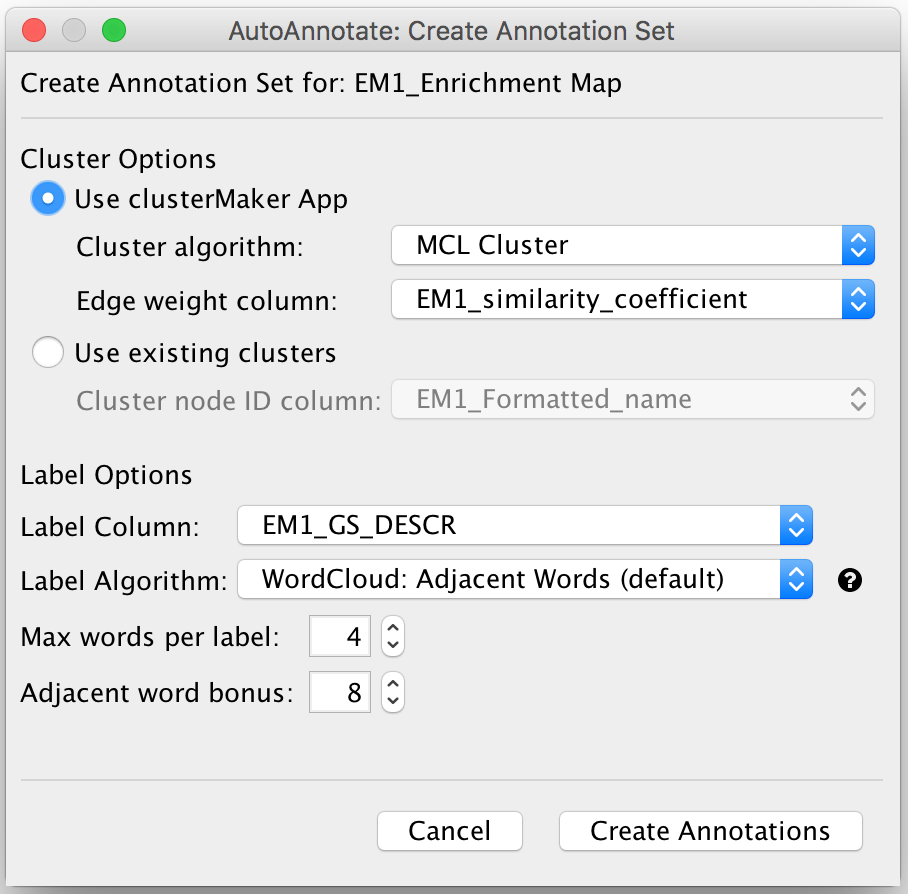
- Cluster Options
- Use clusterMaker App
- If this is selected then clusterMaker will be used to calculate the clusters.
- Cluster algorithm:
- Used to select the clustering algorithm provided by clusterMaker that will be used to calculate the clusters.
See the table below for a list of clusterMaker algorithms currently available for use in AutoAnnotate.
- Edge weight column:
- Some clusterMaker algorithms use edge weights. This box is used to select the Edge Column to use for the edge weights. If the selected algorithm does not use edge weights then this box will be disabled (greyed out).
- User-defined clusters
- You may provide your own clusters or use another clustering app to calculate the clusters. In this case the clusters must already be calculated and cluster identifiers must be stored in a Node Column.
- Any Node Column of discrete type may be used to identify the clusters. (String, Integer, Long Integer, Boolean, or Lists of any of those types.)
- If you select a column of List type then the clusters may overlap.
- Use clusterMaker App
- Label Options
- Label Column
Select a Node Column of type String that will be used to calculate the cluster labels.
- Label Algorithm
- Select an algorithm that is used to calculuate the label for each cluster.
- Two label algorithms are currently available:
WordCloud: Adjacent Words
Uses the WordCloud app to calculate the labels.
Basically the same algorithm that is used by the "Cloud Style: Clustered Standard" option in WordCloud.
- Words in the label are chosen from the most frequent words in the cluster, and words that are adjacent to the most frequent words.
- The higher the "adjacent word bonus" is, the more likely adjacent words will be in the label.
WordCloud: Largest Words
- Label Column
Click Create Annotations to create the new Annotation Set.
WARNING: AutoAnnotate will manage all groups (compound nodes) for the current network view. It is not recommended to manually create groups on a network view that has Annotation Sets. If you would like to manually create groups at any time please duplicate the network view.
ClusterMaker2 Algorithms
The following clusterMaker2 algorithms are made available in AutoAnnotate:
Algorithm |
Weighted? |
Affinity Propagation Cluster |
Yes |
Cluster Fuzzifier |
Yes |
Community cluster (GLay) |
No |
ConnectedComponents Cluster |
Yes |
Fuzzy C-Means Cluster |
Yes |
MCL Cluster |
Yes |
SCPS Cluster |
Yes |
Weighted algorithms require an edge attribute to be used as the edge weight.
Panels
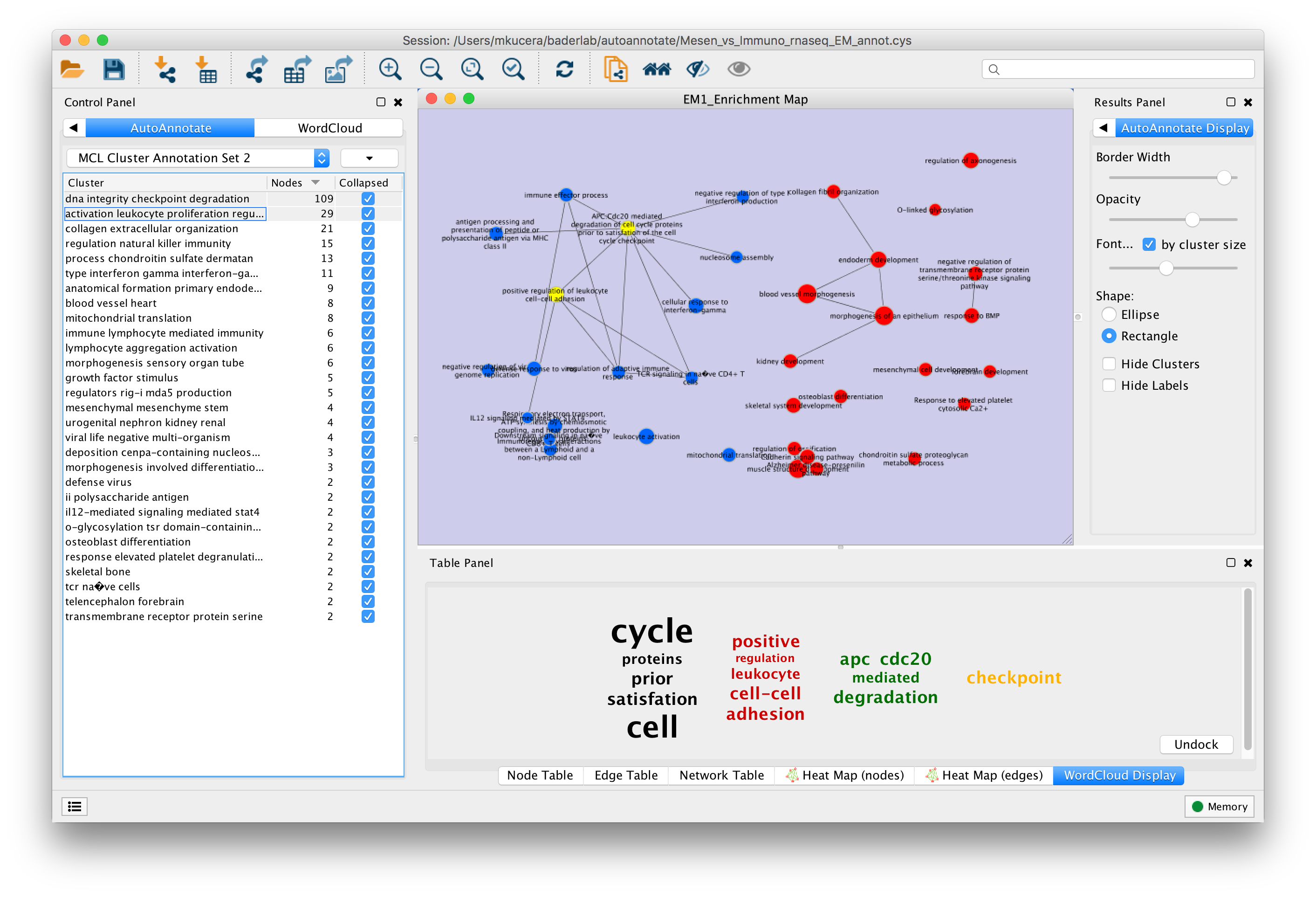
When an Annotation Set is created the AutoAnnotate panels will be shown.
Note: To show or hide the AutoAnnotate panels at any time go to the main menu and select Apps > AutoAnnotate > (Show/Hide) AutoAnnotate.
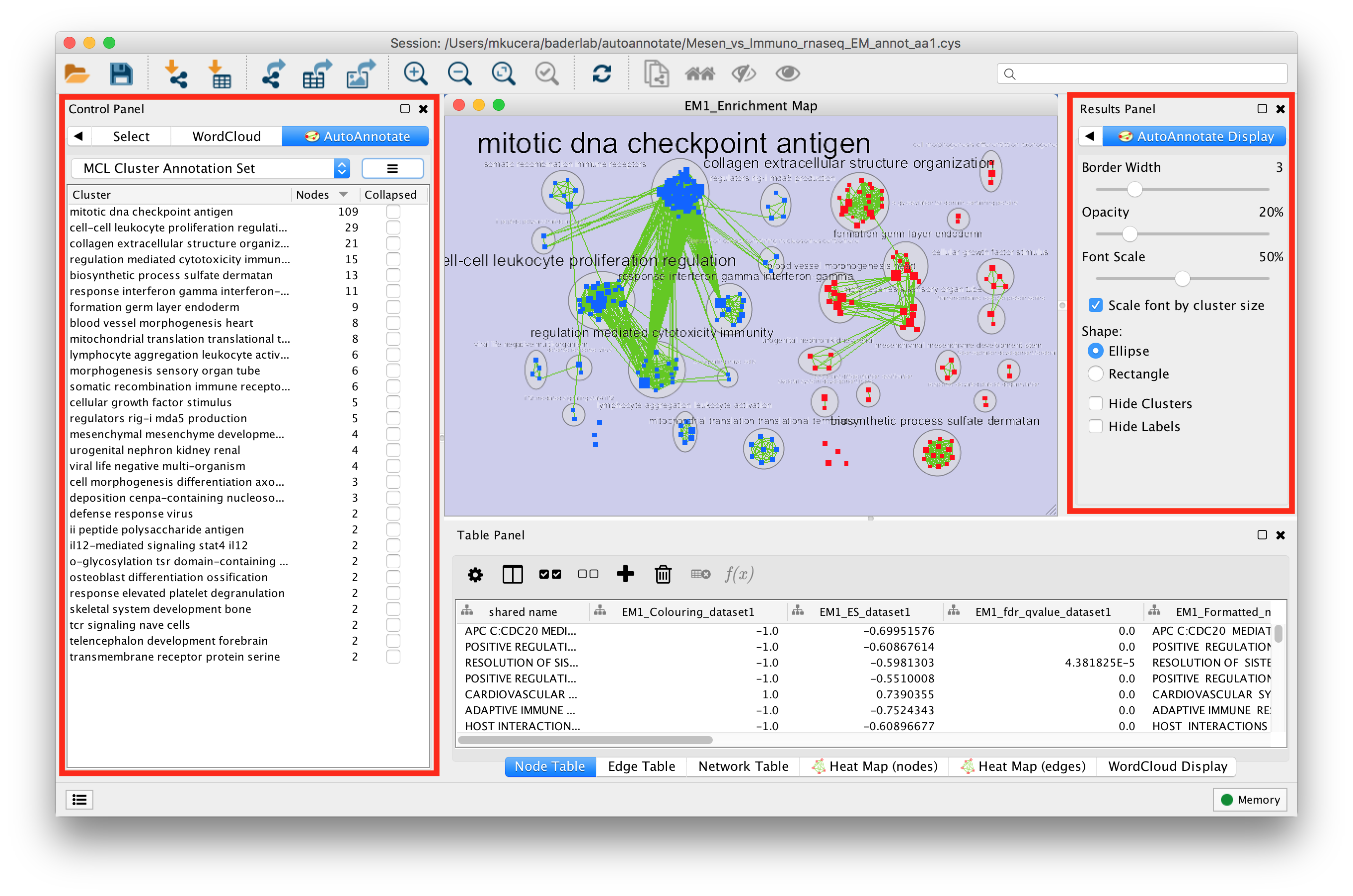
In the Control Panel on the left side is the main AutoAnnotate panel.
- This panel is used to manage Annotation Sets and Clusters.
In the Results Panel on the right side is the AutoAnnotate Display panel.
- This panel is used to modify the look of the annotations.
Changing Display Options
This AutoAnnotate Display panel shows display options for the currently selected Annotation Set.
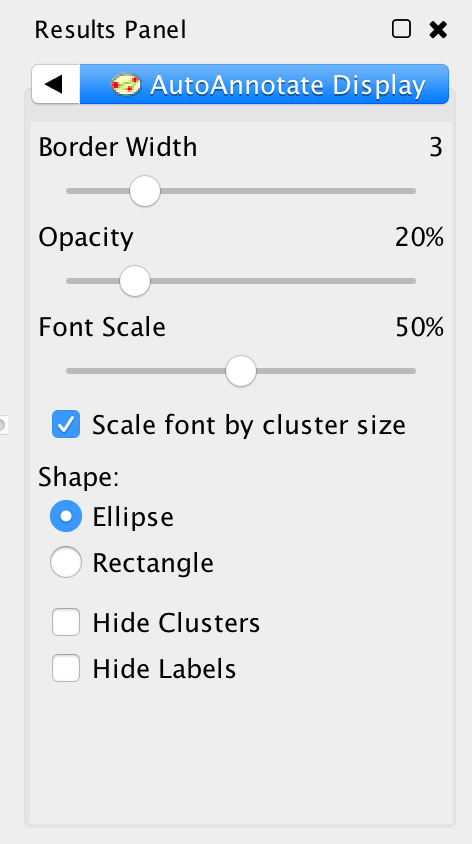
- Border width
- Slider that adjusts the width of the cluster annotation borders.
- Opacity
- Slider that adjusts the opacity of the cluster annotation fill color.
- Slide to the left for completely transparent. Slide to the right for completely opaque.
- Font Size
- Scale font by cluster size
- When selected the size of the label will be relative to the number of nodes in the cluster. Larger clusters will have larger labels.
- When deselected all the clusters will have the same size labels.
- Slider is used to adjust the font size or font scale.
- Scale font by cluster size
- Shape
- There are two available shapes: Ellipse or Rectangle
- Hide Clusters
- Select to hide all cluster shape annotations.
- Hide Labels
- Select to hide all cluster labels.
Managing Annotation Sets
The main AutoAnnotate panel is used to manage Annotation Sets.
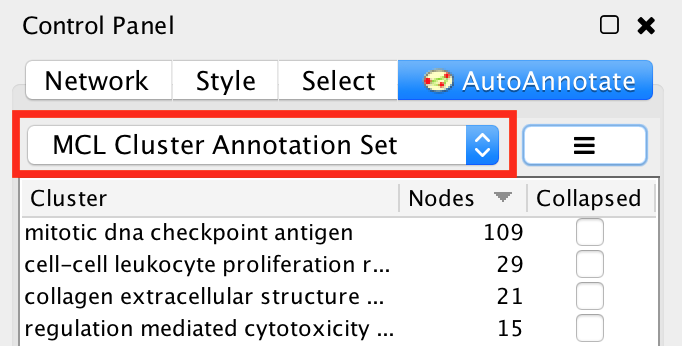
At the top of the panel is a dropdown box that allows you to switch between all of the Annotation Sets that have been created for the current network view.
- Select the (none) option to clear all annotations from the view.
- Select an Annotation Set to show all the annotations and labels in that set.
- When switching between Annotation Sets all the annotations in the view will be erased and redrawn.
- Similarly when switching Annotation Sets any collapsed groups will be expanded.
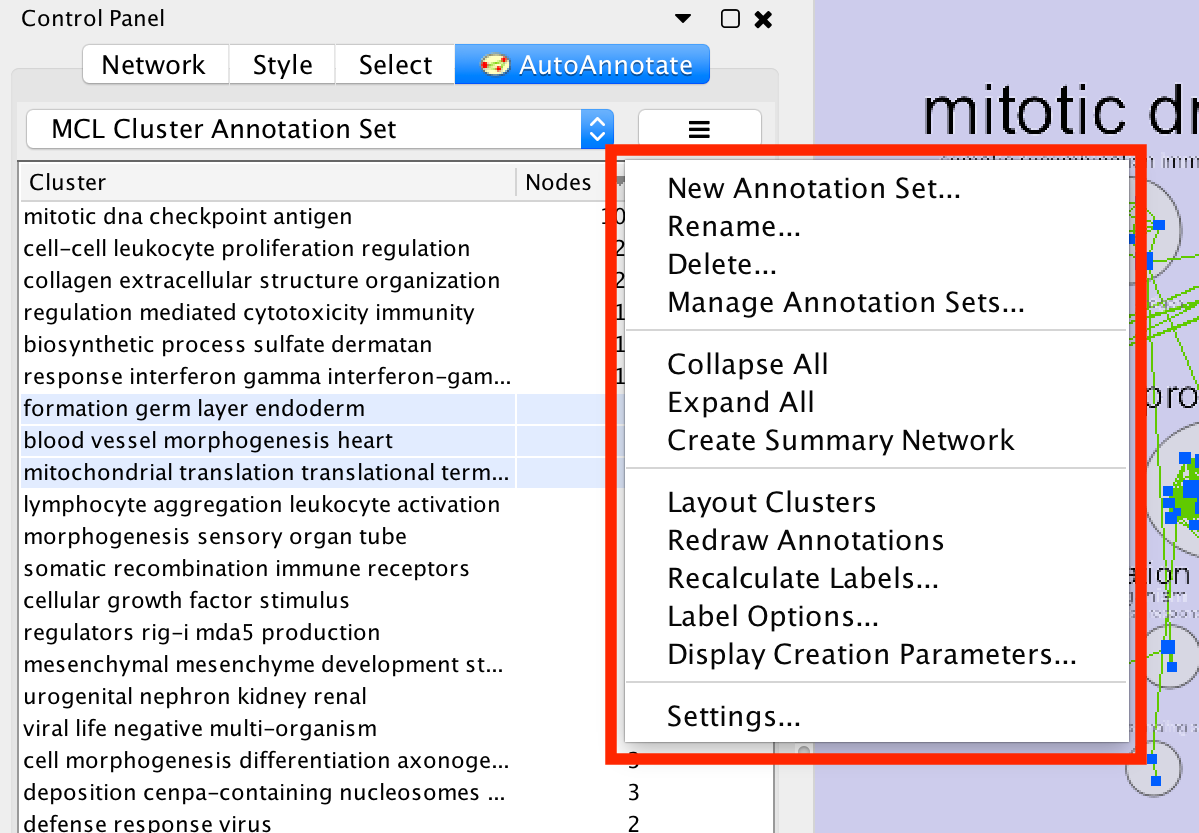
- Next to the dropdown box is the menu button. Click this button to show the Annotation Set Menu.
- New Annotation Set…
- Shows the Create Annotation Set dialog.
- Rename
- Renames the current annotation set
- Delete...
- Deletes the current annotation set
- Manage Annotation Sets...
- Opens a Dialog where you can:
- Change the order that Annotation Sets appear in the dropdown.
- Delete multiple Annotation Sets at a time.
- Opens a Dialog where you can:
- Collapse All
- Collapses all the clusters
- Warning: Collapsing all clusters in a large network can be very slow. Consider using the "Create Summary Network" option instead.
- Expand all
- Expands all the clusters
- Create Summary Network
- Generates a new network where each node represents a collapsed cluster in the original network.
- Very similar in functionality to "Collapse All", but runs significantly faster.
- Layout Clusters
- Runs a layout algorithm that will layout each cluster separately using the perfuse layout (evenly spaced in a grid). Note: this operation cannot be undone.
- Redraw annotations
- Sometimes the annotations may need to be redrawn.
- Recalculate Labels
Recalculates the labels for all of the clusters in the annotation set. Typically used when the WordCloud options or label algorithm changes. More on this later in this guide. (Note: to recalculate labels for a subset of clusters please select the clusters in the table and right-click to show the cluster context menu.)
- Label Options
- Opens a dialog where you can select the label algorithm that is used when recalculating labels.
- Settings...
Opens the AutoAnnotate settings dialog.
- New Annotation Set…
Managing Clusters in the AutoAnnotate Panel
On the main AutoAnnotate panel you will find the Cluster Table.
- This table shows each cluster, along with its label, the number of nodes in the cluster and a checkbox icon that indicates if the cluster has been collapsed.
- Click on a table header to sort the table by that column.
Most of the cluster options are available using the context menu of the cluster table.
- Start by selecting one or more clusters in the cluster table.
- To select multiple clusters hold the Shift key to select a range or hold the Ctrl/Command key to select individual clusters.
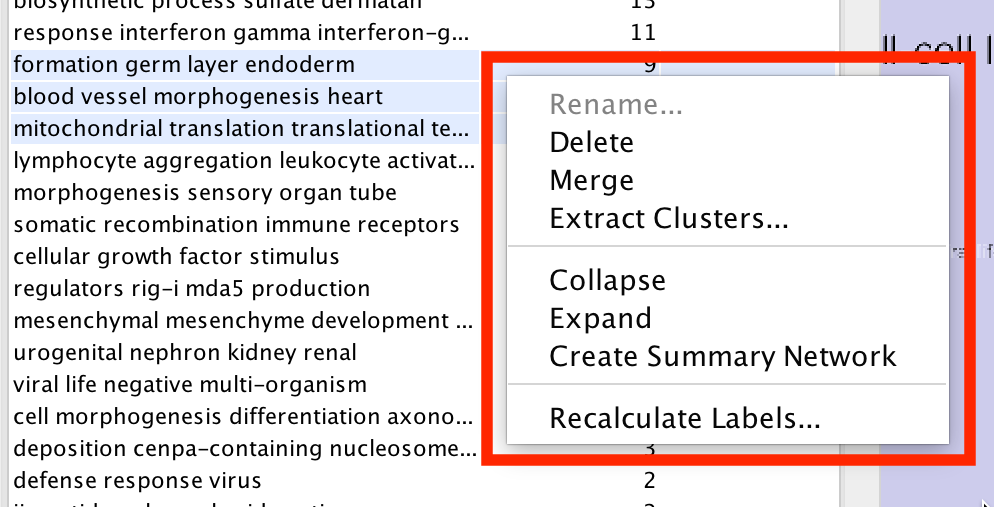
Right click with he mouse to show the cluster table context menu.
- Rename…
- Enabled when 1 cluster is selected.
- Manually set the label for the cluster.
- Delete
- Delete the cluster annotations
- Note: This does not delete the nodes or edges in the cluster, just the annotations.
- Delete the cluster annotations
- Merge
- Enabled when 2 or more clusters are selected.
- Merges the clusters into a single cluster.
- A new label for the merged cluster is automatically calculated.
- Extract Clusters…
- Creates a new Annotation Set consisting of the selected clusters.
- Collapse
- Collapses the selected clusters
- Warning: can be very slow when many clusters are selected.
- Expand
- Expands the selected clusters
- Generate Summary Network
- Generates a new network where each node represents a collapsed cluster that is selected in the original network.
- Very similar in functionality to "Collapse", but runs significantly faster.
- Recalculate Labels
- Recalculates the labels for the selected clusters.
- Rename…
Managing Clusters in the Network View
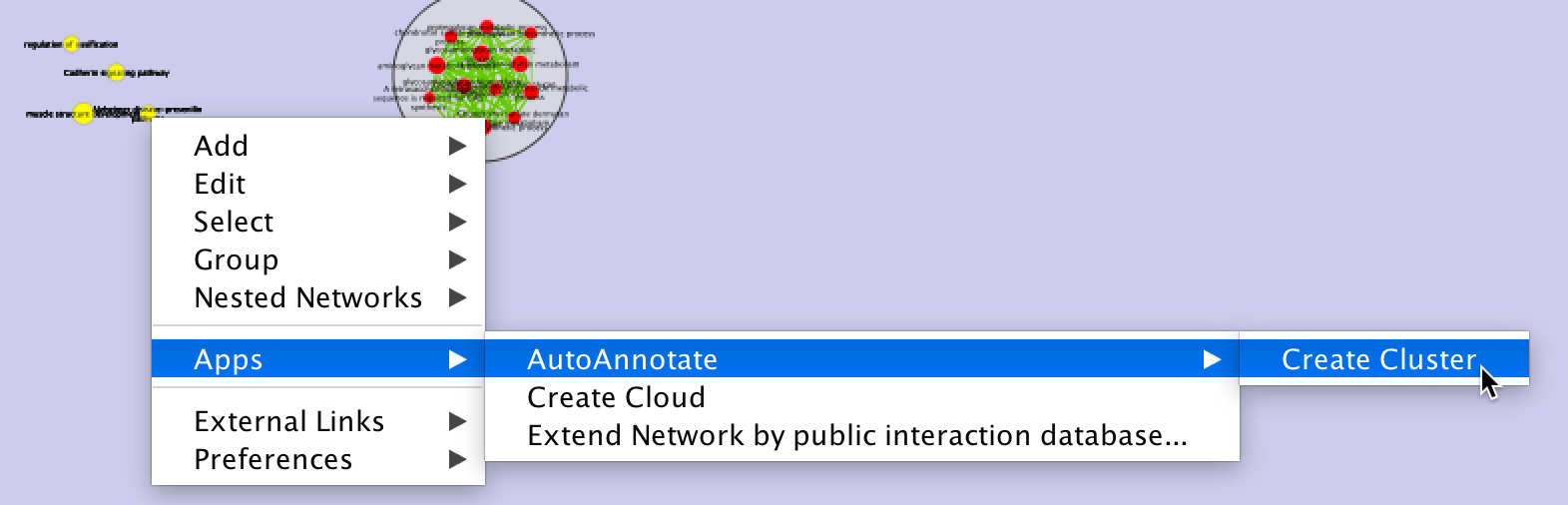
- Manually create a cluster
Select some nodes in the network view, right click to show the context menu, and select Apps > AutoAnnotate > Create Cluster.
- A new cluster is created.
- A label for the cluster is automatically calculated.
- The cluster is selected in the cluster table.
- Deleting a node in an existing cluster will remove the node from the cluster.
- A new label is calculated.
Collapsing and Expanding Clusters
AutoAnnotate can summarize a network by collapsing some or all of the clusters automatically.
Warning, as of Cytoscape 3.4 this function can be very slow, depending on the number of clusters. Consider using the "Generate Summary Network" option instead because it runs significantly faster. See the section below this one for more details.
- The Cytoscape "group nodes" feature is used to collapse and expand clusters. A group node is a compound node that contains other nodes.
When AutoAnnotate collapses a cluster it first creates a group node that contains all the nodes in the cluster and then the group node is collapsed. When the cluster is collapsed Cytoscape will create "meta-edges" between the group node any other nodes it is connected to. The collapsed group nodes and the meta-edges provide a summary of the network.
- The annotation for a cluster is not shown when the cluster is collapsed. The label shown for a collapsed cluster is generated by Cytoscape and depends on the Group Preferences (explained below).
- When a cluster is expanded the group node is expanded and then deleted. The annotations are show again.
It is not recommend to manually collapse or expand clusters using the Groups menu as this may confuse AutoAnnotate.
- When switching between Annotation Sets all collapsed clusters are automatically expanded.
Clusters can be collapsed or expanded from the Annotation Set Menu or the Cluster Table Context Menu.
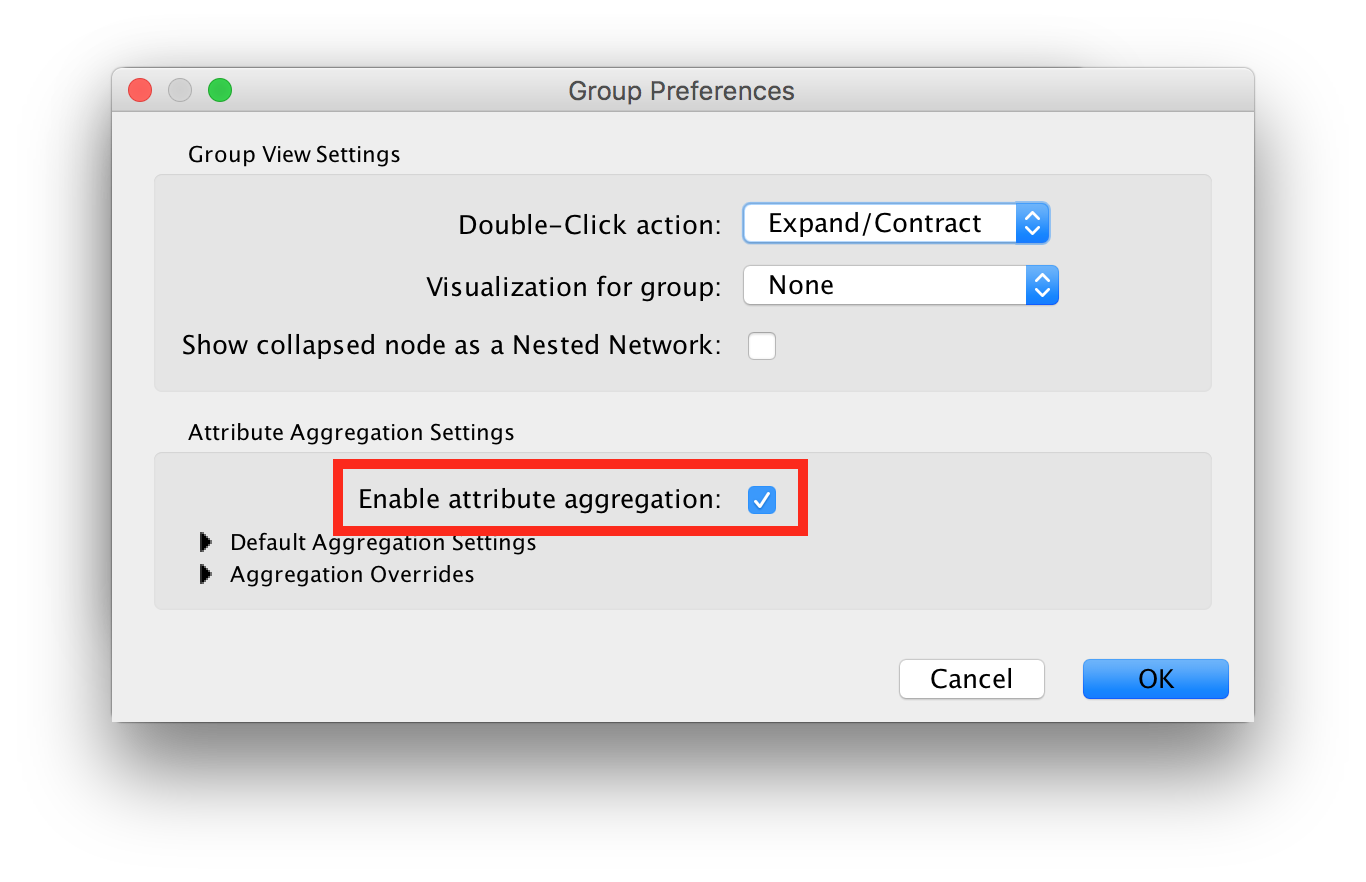
- Before collapsing clusters make sure that node aggregation is enabled. This will tell Cytoscape to generate a label for the group node that aggregates the labels of the nodes in the cluster.
In the main menu select Edit > Preferences > Group Preferences.
In the Group Preferences dialog select the option for Enable Attribute Aggregation.
Summary Network
Using group nodes to collapse clusters can be very slow when there are many clusters. As an alternative a new option called Generate Summary Network has been added in AutoAnnotate 1.1. A new network is generated where each node represents a collapsed cluster in the original network, and each edge represents a meta-edge between clusters. The resulting network is very similar to the results you get from collapsing the clusters.
The main differences are:
- Nodes that are not contained in any cluster are ignored.
- There is no relationship maintained back to the original network. If the clusters in the original network change the summary network will not be updated. Create a new summary network at any time to reflect the current state of the clusters.
- A new visual style for the summary network is created based on the original visual style.
A summary network can be created from the Annotation Set Menu or the Cluster Table Context Menu.
Uses the same attribute aggregation settings as collapsed group nodes. Available from Edit > Preferences > Group Preferences then expand the Default Aggregation Settings section.
Working with WordCloud
Labels for clusters are generated by the WordCloud app.
You may change the label generation options from the WordCloud panel.
In the WordCloud panel you can change the label options, for example using the Normalize slider.
Back in the AutoAnnotate panel select Recalculate Labels to regenerate labels using the updated WordCloud options.
Saving and Restoring a Session with Annotation Sets
- There are currently some limitations in Cytoscape with regards to saving and restoring annotations and groups in a session file.
For this reason when you save your session all the clusters must be expanded first. This is planned to be fixed in future versions of Cytoscape and AutoAnnotate.
Additional Options
AutoAnnotate has various warning dialogs that can be dismissed permanently by selecting "don’t show this again". If you would like to see these warnings again they can be re-enabled. In the main menu select Edit > Preferences > Properties, then select org.baderlab.autoannotate in the Properties Dialog.  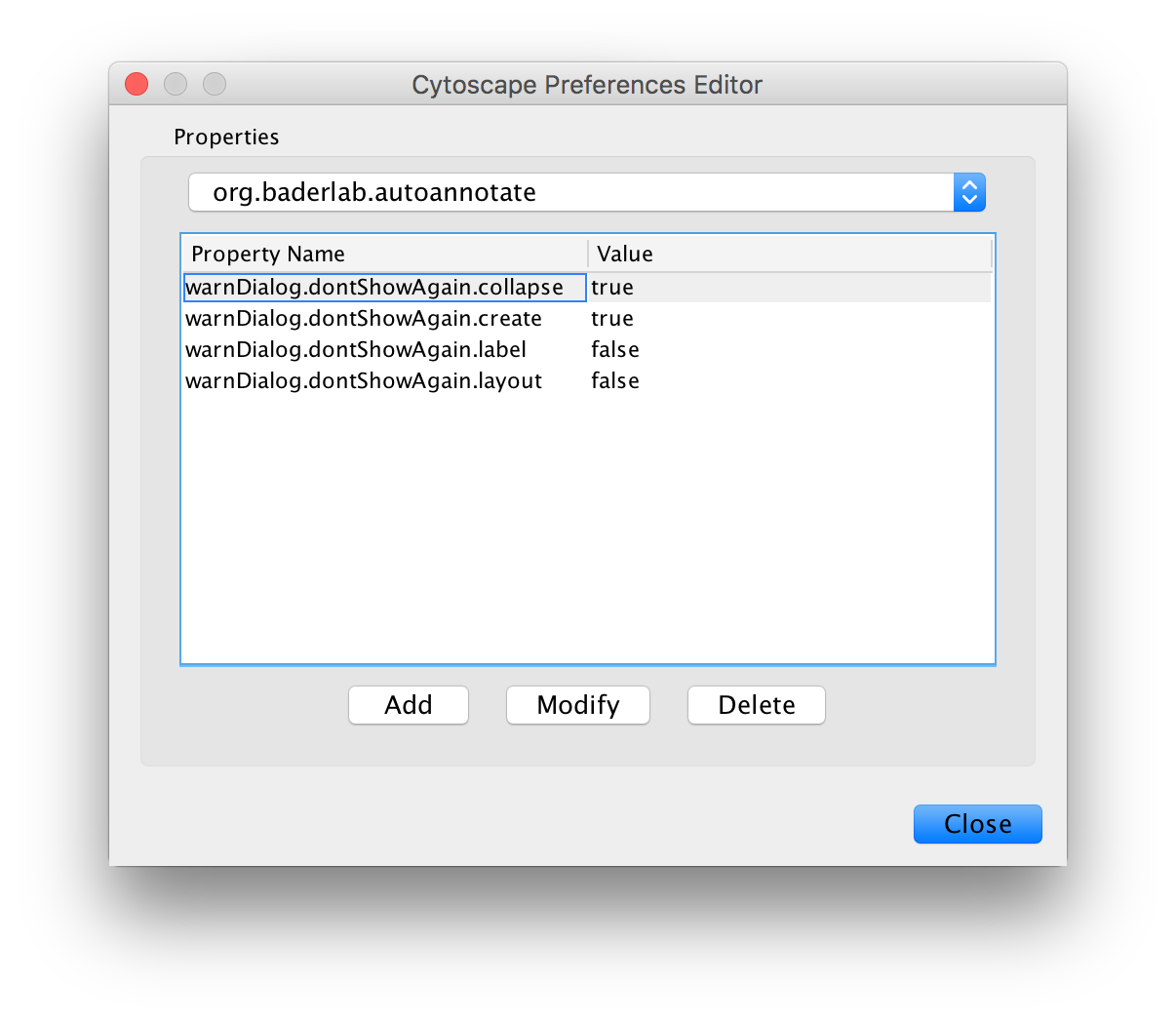
Filing Bug Reports and Requesting Enhancements
Thank you for using AutoAnnotate. Your feedback is important and helps us to improve the App for everyone. If you have any comments, requests for new features or enhancements, or bugs to report, please enter them on the AutoAnnotate github issue tracker.
