Annotating an Enrichment Map
Requirements
Cytoscape version at least 3.2.0. At the time of this writing this has not yet been released. It is scheduled for release in late 2014. Until then, the latest working build (UNSTABLE) can be downloaded from here
WordCloud version at least 2.0.2
ClusterMaker version at least 0.9.3
Creating an annotation set
Once you have created your Enrichment Map, select “Annotate Clusters” from Enrichment Map under the Apps menu at the top of the screen to begin annotating.
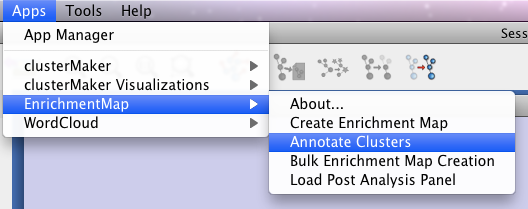
This will open an Annotation Panel in the Cytoscape Control Panel, on the left of the window.
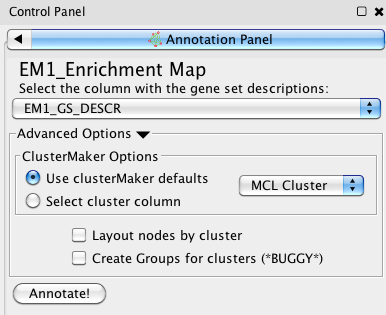
In this panel, you can specify certain parameters for the annotation. Words contained in the gene set description column will be used to automatically generate labels. To proceed immediately, select a clustering algorithm from the dropdown menu. MCL clustering is recommended by default. For more refined clustering, you can run clusterMaker on your own and then specify the column that it creates with your clusters (these will usually look something like 'mclCluster'). You can also specify your own clusters by entering it into a new column and selecting it here. The “Annotate!” button creates a new annotation set. It should be noted that labels are automatically generated using WordCloud, and some manual adjustment will often be necessary.
You will need to select a column from the gene set descriptions drop down menu. This column is used by WordCloud to create a cloud from which the labels for the clusters are generated. This can be any column of strings, but in general labels are generated best when there are more words to work with.
By default, the MCL (Markov Cluster) algorithm is used by clusterMaker. Alternatively, if you want to tune the parameters of clusterMaker yourself, you can run clusterMaker and select the column outputted by clusterMaker by clicking on the “Select cluster column” radio button in the “Advanced Clustering Options panel. These columns must be either integers, or lists of integers.
Selecting the “Layout nodes by cluster” option will rearrange the nodes in your network, grouping them by network and applying a Prefuse Force Directed Layout to each group. This reduces the amount of overlap between clusters and makes the annotations easier to read and interpret. Alternatively, repositioning can be done manually by selecting clusters in the table, dragging the nodes to a new location, and pressing the “Update” button. Multiple clusters can be selected by doing a Command+click for Mac users or Ctrl+click for other operating systems.
The text labels for each cluster can be manually adjusted. It is recommended to look at the WordClouds when doing this, by selecting “WordCloud” in the “Autofocus Preferences” menu. The parameters of WordCloud, such as specifying words to exclude, delimiters, and stemming, can be adjusted in its panel. For these changes to take effect, you must update both the WordCloud and the annotations. You can also manually edit the labels, by double-clicking on the cluster in the table, and changing the text.
How are the labels computed? The labels are computed by gathering all of the descriptions of each gene set and passing them to WordCloud. WordCloud parses these and assigns sizes to each word, proportional to the frequency of the words in the cluster, optionally normalized to consider how frequently the words occur in the entire network. These words are then sorted in order of size. One at a time, the largest word in the cloud is added until the label is 4 words long, or the next largest word is considerably smaller than the most recent word. An advantage is given to words in the same WordCloud groups (colours) as words already added to the label. This advantage can be specified in the Display Options Panel. ‘Considerably smaller’ is defined as below a specified fraction of the previous word’s size - by default set to be 0.3 from the first word to the second, 0.8 from second to third, and 0.9 from third to fourth. This can also be specified in the Display Options Panel. Why are some of the WordClouds blank, but still giving labels? Sometimes, WordCloud assigns a font size of 0 to words appearing infrequently. The annotation process will take this into consideration and still create a label out of these words. Why aren’t all of the clusterMaker algorithms available? Only some of the algorithms in clusterMaker produce output that can be easily parsed and used to partition the nodes for annotations.
Arkady Arkhangorodsky (aarkhangorodsky@gmail.com) Ruth Isserlin (ruth.isserlin@utoronto.ca)
ClusterMaker doesn’t run properly on a second enrichment map Specifying gene set descriptions
Clustering
Laying out clusters
Changing labels
Frequency Asked Questions
Contact
Known bugs