collapse_ExpressionMatrix.py
This tool can process a gene expression matrix (in GCT or TXT format) ranked list (RNK format) and:
convert the Identifier based on a Chip Annotation file (e.g. AffyID -> Gene Symbol)
- collapse the expression values or rank-scores for Genes from more than one probe set.
This can be done either individually or both at the same time.
Requirements
collapse_ExpressionMatrix.py requires:
- Python 2.3 or newer (but not Python 3.x!)
the Tkinter Library (comes with most Python installations) for the GUI
Supported Operating Systems:
MacOS X 10.5 "Leopard" ore newer (probably also MacOS X 10.4 "Tiger")
Windows (download and install the most recent version of Python 2.x from: http://www.python.org/download/ or http://www.activestate.com/activepython/downloads/
Linux (Python and Tcl/Tk are probably already installed out of the box, otherwise install the packages with your Distribution's package manager)
GUI Mode
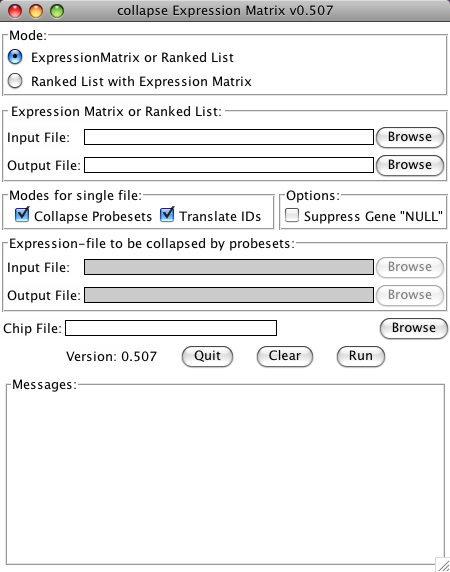 collapse_ExpressionMatrix.py now has a Tk-based Graphical User Interface (GUI). To use the GUI, just start the program without any arguments. This can be done:
collapse_ExpressionMatrix.py now has a Tk-based Graphical User Interface (GUI). To use the GUI, just start the program without any arguments. This can be done:
on Windows: double-click on the collapse_ExpressionMatrix.py-file
- on MacOS 10.5 or newer with installed "Developer Tools":
control-click (or right-click) on the collapse_ExpressionMatrix.py-file in the finder and choose "Open With/Build Applet.app"
this will create an MacOS Application collapse_ExpressionMatrix.app which can be started by double clicking.
- on MacOS, Linux or other Unix-like Systems in a Terminal/Shell: see in section "Command Line Mode" how to make the program executable.
After starting the GUI:
- use the first Browse-Button to select an Expression Matrix or Ranked gene list as an input file.
- use the second Browse-Button to choose a name and location of the output file (the program will suggest to use the same name as the input file with an inserted "_collapsed" before the extension)
- choose if the Identifiers should be converted or the file should be collapsed by checking the check-boxes
- if Identifiers are to be converted, choose a matching chip file
- start the conversion by clicking the Run button
Command Line Mode
If you are familiar with command line tools under Unix/Linux, collapse_ExpressionMatrix.py -h gives you all the information you need (if not, see below):
$ collapse_ExpressionMatrix.py -h
Usage: collapse_ExpressionMatrix_SVN.py [options] -i input.gct -o output.gct [-c platform.chip] [--collapse]
This tool can process a gene expression matrix (in GCT or TXT format) or
ranked list (RNK format) and either replace the Identifier based on a Chip
Annotation file (e.g. AffyID -> Gene Symbol), or collapse the expression
values or rank-scores for Genes from more than one probe set. Both can be done
in one step by using both '-c platform.chip' and '--collapse' at the same
time. If a ranked list is to be collapsed, an additional expression matrix can
be supplied by the -e/-x parameters and will be filtered to contain the same
probe-sets as selected from the RNK file. If however the file supplied by -i
is not recognized as a RNK file, these options have no effect. For detailed
descriptions of the file formats, please refer to:
http://www.broadinstitute.org/cancer/software/gsea/wiki/index.php/Data_formats
Call without any parameters to select the files and options with a GUI
(Graphical User Interface)
Options:
--version show program's version number and exit
-h, --help show this help message and exit
-i FILE, --input=FILE
input expression table or ranked list
-o FILE, --output=FILE
output expression table or ranked list
-c FILE, --chip=FILE Chip File This implies that the Identifiers are to be
replaced.
-e FILE, --ei=FILE (optional) additional input Expression-table, to be
restricted to the same probe-sets as the RNK file
-x FILE, --xo=FILE (optional) corresponding output file for -i/--ei
option
--collapse Collapse multiple probe sets for the same gene symbol
(max_probe)
--no-collapse Don't collapse multiple probesets [default]
--null suppress Gene with Symbol NULL
-g, --gui Open a Window to choose the files and options.
-q, --quiet be quiet
On MacOS and Linux you need to make the program executable. Therefore:
copy the file to a directory, e.g. ${HOME}/bin
- open a Terminal
set the eXecutable flag:
chmod a+x ${HOME}/bin/collapse_ExpressionMatrix.pyif the ${HOME}/bin directory is not in your search Path (test by running collapse_ExpressionMatrix.py from a terminal) add it by adding the line export PATH=$HOME/bin:$PATH to your ${HOME}/.bash_profile using your favourite text editor (pico, vi, emacs, gedit, TextWrangler, etc.) or with the command
echo export PATH=\$HOME/bin:\$PATH >> ${HOME}/.bash_profile, or refer to your local SysAdmin for any other shell that bash.
open a new terminal or run source ${HOME}/.bash_profile
test with collapse_ExpressionMatrix.py -h
On Windows:
copy the file to a directory, e.g. C:\bin
- open the Conrtol Panel
- open System
- go to Advanced System Settings (on Vista and 7 only)
- go to the Advanced Tab
- Click on Environment-button
- if in the section "User variables for {USERNAME}" there is already an entry called "PATH":
- click on Edit...
append ;C:\bin at the very end
- otherwise click on New...
Variable Name: PATH
Variable Value: %PATH%;C:\bin
open a Command Prompt (Programs/Accessories)
test with collapse_ExpressionMatrix.py -h