|
Size: 2284
Comment:
|
Size: 2442
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 30: | Line 30: |
| * SigGMT / Expression: `Estrogen_expression_file.txt` (OPTIONAL) | * SigGMT / 'CTCF_DIFF.gmt' |
| Line 34: | Line 34: |
| * in the 'Signature-Genesets' box: click on CTCF_DIFF as available Signature-Genesets | * in the '''Signature-Genesets''' box: click on CTCF_DIFF as available Signature-Genesets |
| Line 39: | Line 39: |
| * Set 'Number of common genes' to 20 * Click on "Run" |
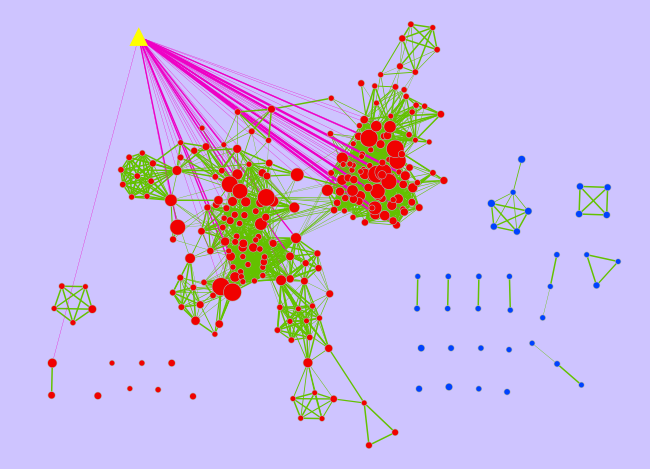
* Set '''Number of common genes''' to 20 *6. Click on '''Run''' {{attachment:PostAnalysis_screenshot.png}} <<BR>><<BR>> == Examining Results == [[attachment:ES12_EM_example_PA_CTCF_Diff.cys]] {{attachment:PostAnalysisMap_screenshot.png}} |
Enrichment Map Post Analysis Tutorial
Outline
This quick tutorial will guide you through the creation of an additional gene-set on an pre-existing Enrichment Map. It can for example help to localize microRNA, transcription factors or drug targets in enriched pathways displayed on the map. The new gene-set that we want to add to the network is called the signature gene-set.
To run this tutorial:
- You need to have Cytoscape installed : minimally 2.6.3 must be installed but preferable to have the latest version of Cytoscape 2 (e.g. 2.8.3)
- It does not work with Cytoscape 3
- Install the Enrichment Map plugin from the Cytoscape plugin manager. If you install it manually (e.g. if you need to install a new version that doesn't happen to be in the plugin manager yet), then it must be in the Cytoscape-[Version#]/plugins folder
You need to download the test data: PostAnalysisTutorial.zip
Description of the tutorial files contained in the PostAnalysisTutorial folder:
- CTCF_DIFF.gmt : List of the genes included in the signature gene-set.
Human_GO_AllPathways_no_GO_iea_April_15_2013_symbol.gmt: Original gene-set file that has been used to create the original Enrichment Map
- ES12_EM_example.cys: the Enrichment Map on which we want to add the signature gene-set
Instructions
1. Open Cytoscape
1. Open ES12_EM_example.cys
2. Click on Plugins / Enrichment Map/ Post Analysis
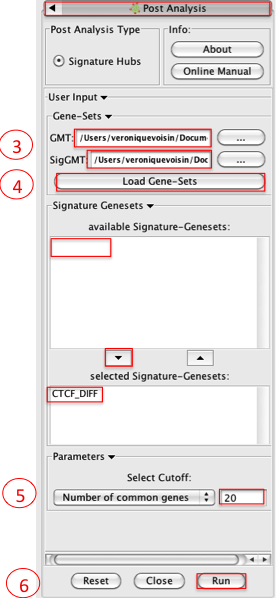
3. Please select the following files by clicking on the respective (...) button and selecting the file in the Dialog:
GMT / 'Human_GO_AllPathways_no_GO_iea_April_15_2013_symbol.gmt'
- SigGMT / 'CTCF_DIFF.gmt'
4. Click on Load Gene-Sets
in the Signature-Genesets box: click on CTCF_DIFF as available Signature-Genesets
- click on the down arrow to move CTCF_DIFF in the lower box
5. Tune parameters
Set Number of common genes to 20
6. Click on Run