|
Size: 6174
Comment:
|
Size: 8028
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| #acl RuthIsserlin:read,write,delete,revert All: | ## page was renamed from Software/EnrichmentMap/GSEATutorial #acl All:read |
| Line 8: | Line 9: |
| This quick tutorial will guide you through the generation of an Enrichment Map for an analysis performed using [[http://www.broadinstitute.org/gsea/| GSEA Gene Set Enrichment Analysis]], | This quick tutorial will guide you through the generation of an Enrichment Map for an analysis performed using [[http://www.broadinstitute.org/gsea/| GSEA Gene Set Enrichment Analysis]]. |
| Line 10: | Line 11: |
| To run this tutorial: * You need to have Cytoscape installed : minimally 2.6.3 must be installed but preferable to have the latest version of cytoscape |
== To run this tutorial == * You need to have Cytoscape installed : minimally 2.6.3 must be installed but preferable to have the latest version of Cytoscape 2 (e.g. 2.8.3) * It does not work with Cytoscape 3 |
| Line 15: | Line 17: |
| Description of the tutorial files contained in the !GSEATutorial folder: * EX_NT.cls : phenotype definition for expression file required by GSEA. |
== Description of the tutorial files contained in the GSEATutorial folder == * ES_NT.cls : phenotype definition for expression file required by GSEA. |
| Line 19: | Line 21: |
| Line 27: | Line 28: |
| * Click on “Save Taget as…” and save shortcut to your desktop or your folder of choice so you can launch GSEA for your analysis without having to navigate to it through your web browser. | * Click on “Save Target as…” and save shortcut to your desktop or your folder of choice so you can launch GSEA for your analysis without having to navigate to it through your web browser. |
| Line 30: | Line 31: |
| 1. Click on ''Browse for files…'' in newly opened '''Load data''' pane. | 1. Click on ''Browse for files…'' in newly opened '''Load data''' panel. |
| Line 34: | Line 35: |
| 1. Click on the arrow next to the ''Expression dataset'' text box. Select the expression set you wish to run the analysis on. 1. Click on ''…'' next to the text box of ''Gene Set Database.'' 1. Click on ''Gene Matrix (local gmx/gmt)'' tab. 1. Select gmt file Human_GO_AllPathways_no_GO_iea_April_15_2013_symbo.gmt and click on ''Ok''. 1. Click on ''…'' next to the text box of ''Phenotype labels''. 1. Make sure ''Select source file'' is set to ES_NT.cls. 1. Select ''ES12_versus_NT12'' and click on ''Ok''. |
1. Select the ''Expression dataset:'' * Click on the arrow next to the ''Expression dataset'' text box. * Select the expression set you wish to run the analysis on (MCF7_ExprMx_v2_names.gct). 1. Select the ''Gene Set Database'': * Click on ''…'' next to the text box of ''Gene Set Database.'' * Click on ''Gene Matrix (local gmx/gmt)'' tab. * Select gmt file Human_GO_AllPathways_no_GO_iea_April_15_2013_symbo.gmt and click on ''Ok''. 1. Select the ''Phenotype labels'' file * Click on ''…'' next to the text box of ''Phenotype labels''. * Make sure ''Select source file'' is set to ES_NT.cls. * Select ''ES12_versus_NT12'' and click on ''Ok''. |
| Line 48: | Line 56: |
| '''Note''': repeat these steps for the 24hrs time-point by repeating steps 14 - end but use ES24_versus_NT24 phenotype labels instead. | '''Note''': repeat steps for the 24hrs time-point but use ES24_versus_NT24 phenotype labels in step 11 instead and in step 15 change the Analysis name (example:estrogen_treatment_24hr_gsea_enrichment_results).. |
| Line 50: | Line 58: |
| === Stpe 2: Generate Enrichment Map with GSEA Output === {{attachment:David_inputpanel.png|Screenshot David Input Panel|align="right"}} |
=== Step 2: Generate Enrichment Map with GSEA Output === {{attachment:GSEA_inputpanel.png|Screenshot David Input Panel|align="right"}} |
| Line 55: | Line 63: |
| 1. Please select the following files by clicking on the respective (...) button and selecting the file in the Dialog: * '''NO GMT file is required for DAVID Analysis''' * Dataset 1 / Expression: `Estrogen_expression_file.txt` (OPTIONAL) * Dataset 1 / Enrichments: `12hr_David_Output.txt` * Click on "''Dataset 2 {{attachment:arrow_collapsed.gif}}''" to expand the panel * Dataset 2 / Expression: ''leave empty'' * Dataset 2 / Enrichments 1: `24hr_David_Output.txt` (OPTIONAL) |
1. '''OPTION 1 - Manually load all files''' Please select the following files by clicking on the respective (...) button and selecting the file in the Dialog: * Gene Sets / GMT : `Human_GO_AllPathways_no_GO_iea_April_15_2013_symbol.gmt` (can be found in directory where you extracted the files downloaded in GSEATutorial.zip) * Dataset 1 / Expression: `MCF7_ExprMx_v2_names.gct` (can be found in directory where you extracted the files downloaded in GSEATutorial.zip) * Dataset 1 / Enrichments 1: `gsea_report_for_ES12_#############.xls` (can be found in directory where you put the GSEA results specified in Part 1, step 15) * Dataset 1 / Enrichments 2: `gsea_report_for_NT12_#############.xls` (can be found in directory where you put the GSEA results specified in Part 1, step 15) * Click on "''Advanced {{attachment:arrow_collapsed.gif}}''" to expand the panel * Dataset 1 / Ranks: `ranked_gene_list_ES12_versus_NT12_1367261038781.xls` (OPTIONAL) (can be found in directory where you put the GSEA results specified in Part 1, step 15) * Dataset 1 / Phenotypes 1: `ES12` VS `NT12` (OPTIONAL) 1. '''OPTION 2 - Populate all fields using GSEA rpt file''' * Dataset 1 / Expression : `ES12vsNT12.Gsea.#############.rpt` (can be found in directory where you put the GSEA results specified in Part 1, step 15) * NOTE: If you are populating the fields using a rpt file and any of the file names appear in red font then the file EM needs was not found. This can happen if you move your GSEA results folders around after they have been created. For the missing file follow step 5 and re-populate the effected fields. |
| Line 66: | Line 81: |
| * Overlap coefficient cut-off `0.6` | * Overlap coefficient cut-off `0.5` |
| Line 70: | Line 85: |
| <<BR>><<BR>><<BR>><<BR>><<BR>><<BR>><<BR>><<BR>> | <<BR>> |
| Line 72: | Line 90: |
| * Example EM session - Estrogen treatment vs no treatment at 12hr [[attachment:ES12_EM_example.cys]] |
|
| Line 75: | Line 95: |
| 1. Node border (outer circle) size corresponds to the number of genes in dataset 2 within the geneset 1. Colour of the node (inner circle) and border(outer circle) corresponds to the significance of the geneset for dataset 1 and dataset 2, respectively. |
1. Colour of the node (inner circle) corresponds to the significance of the geneset for dataset 1. |
| Line 78: | Line 97: |
| * '''NOTE''': if you are using two enrichment sets you will see two different colours of edges in the enrichment map. When the set of genes in the two datasets are different (for example, when you are comparing two different species or when you are comparing results from two different platforms) the overlaps are computed for each dataset separately as there is a different set of genes that the enrichments were calculated on. In this case, since the enrichments were reduced to only a subset of most differentially expressed at each time point the set of genes the enrichments are calculated on are different and overlap are calculated for each set separately. |
'''GSEA Leading Edge Information''': 1. Click on a node (gene set) in the Enrichment map. 1. In the Data Panel, expression profile of all genes included in the selected gene-set should appear in the '''EM GenesetExpression''' viewer tab 1. Change the Normalization to your desired metric. 1. Change the Sorting method to ''GSEARanking''. 1. Genes part of the leading edge are highlighted in yellow. <<BR>> {{attachment:GSEA_leadingedge.png|GSEA EM leading edge}} *'''Note''': Leading edge information is currently only available when looking at a single gene set == For more detailed tutorials check out: == * [[http://www.ncbi.nlm.nih.gov/pubmed/21877285|Visualizing gene-set enrichment results using the Cytoscape plug-in enrichment map. Merico D, Isserlin R, Bader GD. Methods Mol Biol. 2011;781:257-77. doi: 10.1007/978-1-61779-276-2_12.]] * [[http://www.ncbi.nlm.nih.gov/pubmed/23606248|Global proteomic profiling and enrichment maps of dilated cardiomyopathy. Isserlin R, Merico D, Emili A. Methods Mol Biol. 2013;1005:53-66. doi: 10.1007/978-1-62703-386-2_5.]] |
Enrichment Map GSEA Tutorial
Contents
Outline
This quick tutorial will guide you through the generation of an Enrichment Map for an analysis performed using GSEA Gene Set Enrichment Analysis.
To run this tutorial
- You need to have Cytoscape installed : minimally 2.6.3 must be installed but preferable to have the latest version of Cytoscape 2 (e.g. 2.8.3)
- It does not work with Cytoscape 3
- Install the Enrichment Map plugin from the Cytoscape plugin manager. If you install it manually (e.g. if you need to install a new version that doesn't happen to be in the plugin manager yet), then it must be in the Cytoscape-[Version#]/plugins folder
You need to download the test data: GSEATutorial.zip
Description of the tutorial files contained in the GSEATutorial folder
- ES_NT.cls : phenotype definition for expression file required by GSEA.
- MCF_ExpMX_v2_names.gct : Expression File - Estrogen treatment, Official Gene Name as key. - Data for 12hr,24hr and 48hr.
Human_GO_AllPathways_no_GO_iea_April_15_2013_symbol.gmt: Gene set definition file.
Instructions
Step 1: Generate GSEA output files
GO to GSEA website - http://www.broadinstitute.org/gsea/
- Click on Downloads in the page header.
From the javaGSEA Desktop Application right click on Launch with 1 Gb memory.
- Click on “Save Target as…” and save shortcut to your desktop or your folder of choice so you can launch GSEA for your analysis without having to navigate to it through your web browser.
- Double click on GSEA icon you created.
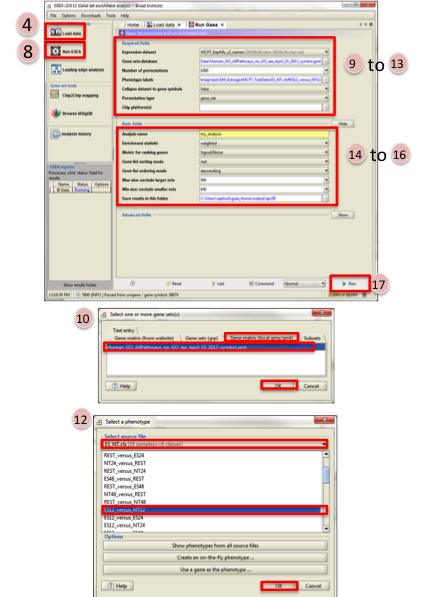
Click on Load data in left panel.
Click on Browse for files… in newly opened Load data panel.
Navigate to directory where you stored tutorial test set files. Select raw expression (.gct) file, sample class file(.cls) and gene set (.gmt) file. Click on Open.
Wait until confirmation box appears indicating that all files loaded successfully. Click on Ok.
Click on Run GSEA in left panel.
Select the Expression dataset:
Click on the arrow next to the Expression dataset text box.
Select the expression set you wish to run the analysis on (MCF7_ExprMx_v2_names.gct).
Select the Gene Set Database:
Click on … next to the text box of Gene Set Database.
Click on Gene Matrix (local gmx/gmt) tab.
Select gmt file Human_GO_AllPathways_no_GO_iea_April_15_2013_symbo.gmt and click on Ok.
Select the Phenotype labels file
Click on … next to the text box of Phenotype labels.
Make sure Select source file is set to ES_NT.cls.
Select ES12_versus_NT12 and click on Ok.
Click on the down arrow next to the text box for Collapse dataset to gene symbols. Select false.
Click on the down arrow next to the text box for Permutation type. Select gene_set.
Click on Show next to Basic fields.
Click in text box next to Analysis name and rename (example:estrogen_treatment_12hr_gsea_enrichment_results).
Click on … next to “Save results in this folder text box. Navigate to the folder where you wish to save the results (preferably the same directory where all the input files have been saved).
Click on Run in the bottom right corner.
Note: repeat steps for the 24hrs time-point but use ES24_versus_NT24 phenotype labels in step 11 instead and in step 15 change the Analysis name (example:estrogen_treatment_24hr_gsea_enrichment_results)..
Step 2: Generate Enrichment Map with GSEA Output
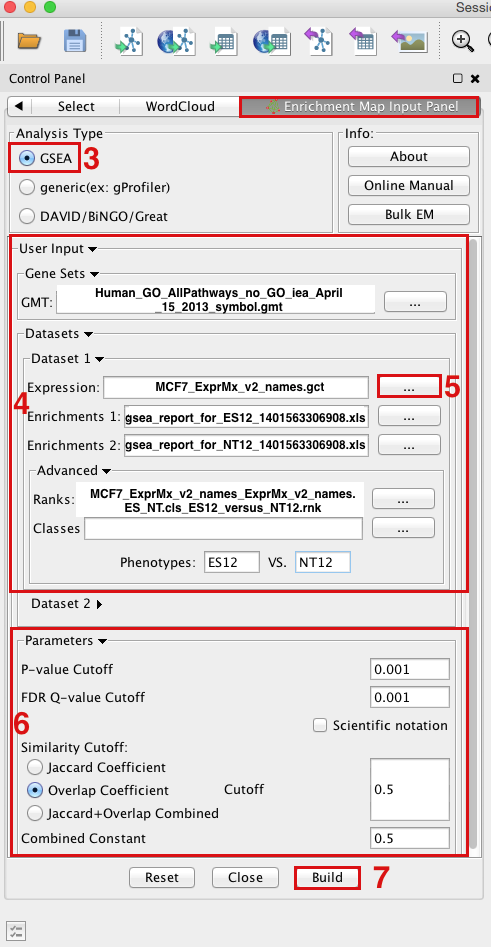
- Open Cytoscape
- Click on Plugins / Enrichment Maps / Load Enrichment Results
- Make sure the Analysis Type is set to GSEA
OPTION 1 - Manually load all files Please select the following files by clicking on the respective (...) button and selecting the file in the Dialog:
Gene Sets / GMT : Human_GO_AllPathways_no_GO_iea_April_15_2013_symbol.gmt (can be found in directory where you extracted the files downloaded in GSEATutorial.zip)
Dataset 1 / Expression: MCF7_ExprMx_v2_names.gct (can be found in directory where you extracted the files downloaded in GSEATutorial.zip)
Dataset 1 / Enrichments 1: gsea_report_for_ES12_#############.xls (can be found in directory where you put the GSEA results specified in Part 1, step 15)
Dataset 1 / Enrichments 2: gsea_report_for_NT12_#############.xls (can be found in directory where you put the GSEA results specified in Part 1, step 15)
Click on "Advanced
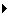 " to expand the panel
" to expand the panel Dataset 1 / Ranks: ranked_gene_list_ES12_versus_NT12_1367261038781.xls (OPTIONAL) (can be found in directory where you put the GSEA results specified in Part 1, step 15)
Dataset 1 / Phenotypes 1: ES12 VS NT12 (OPTIONAL)
OPTION 2 - Populate all fields using GSEA rpt file
Dataset 1 / Expression : ES12vsNT12.Gsea.#############.rpt (can be found in directory where you put the GSEA results specified in Part 1, step 15)
- NOTE: If you are populating the fields using a rpt file and any of the file names appear in red font then the file EM needs was not found. This can happen if you move your GSEA results folders around after they have been created. For the missing file follow step 5 and re-populate the effected fields.
- Tune Parameters
P-value cut-off 0.001
Q-value cut-off 0.05
- Check Overlap Coefficient
Overlap coefficient cut-off 0.5
- Build Enrichment Map
- Go to View, and activate Show Graphics Details
Step 3: Examining Results
Example EM session - Estrogen treatment vs no treatment at 12hr ES12_EM_example.cys
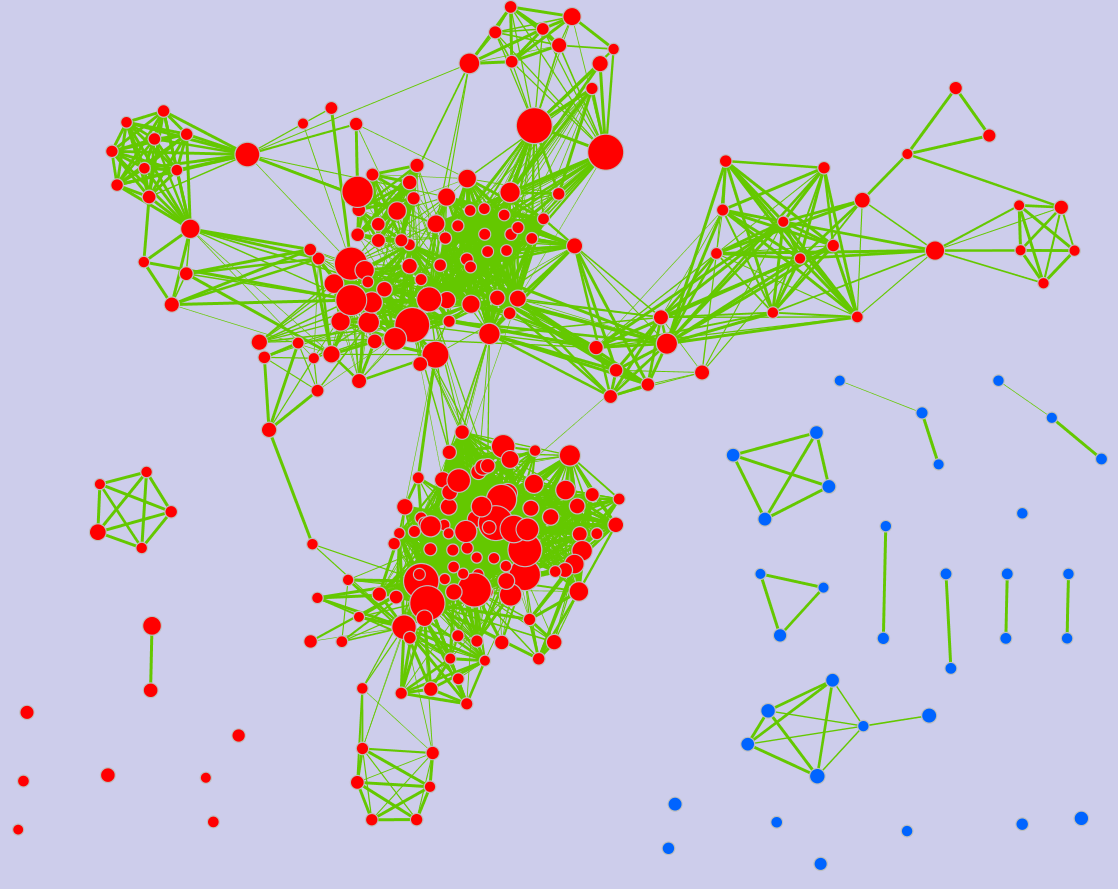
Legend:
- Node (inner circle) size corresponds to the number of genes in dataset 1 within the geneset
- Colour of the node (inner circle) corresponds to the significance of the geneset for dataset 1.
- Edge size corresponds to the number of genes that overlap between the two connected genesets. Green edges correspond to both datasets when it is the only colour edge. When there are two different edge colours, green corresponds to dataset 1 and blue corresponds to dataset 2.
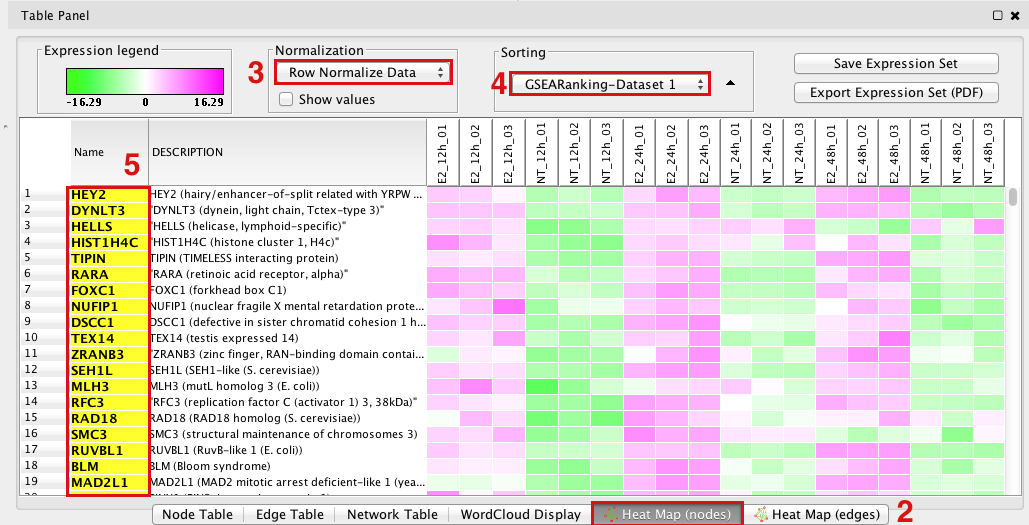
GSEA Leading Edge Information:
- Click on a node (gene set) in the Enrichment map.
In the Data Panel, expression profile of all genes included in the selected gene-set should appear in the EM GenesetExpression viewer tab
- Change the Normalization to your desired metric.
Change the Sorting method to GSEARanking.
- Genes part of the leading edge are highlighted in yellow.
Note: Leading edge information is currently only available when looking at a single gene set
