Enrichment Map g:Profiler Tutorial
Contents
Outline
This quick tutorial will guide you through the generation of an Enrichment Map for an analysis performed using g:Profiler (Functional Profiling of Gene List from large-scale experiments)
To run this tutorial:
- You need to have Cytoscape installed : minimally 2.6.3 must be installed but preferable to have the latest version of Cytoscape (e.g. version 3.1)
Install the Enrichment Map plugin from the Cytoscape plugin manager. If you install it manually (e.g. if you need to install a new version that doesn't happen to be in the plugin manager yet), then it must be in the Cytoscape-[Version#]/plugins folder --> For Cytoscape 2.8
Install the Enrichment Map App from the Cytoscape App manager in cytoscape 3. --> For Cytoscape 3.
Instructions
Step 1: Generate g:Profiler output files
You need to download the test data: gProfilerTutorial.zip
- Description of the tutorial files contained in the gProfilerTutorial folder:
- 12hr_topgenes.txt : List of top genes expressed in Estrogen dataset at 12hr - Official Gene Symbol.
- 24hr_topgenes.txt : List of top genes expressed in Estrogen dataset at 24hr - Official Gene Symbol.
Go to g:Profiler website - http://biit.cs.ut.ee/gprofiler/
Select and copy all genes in the tutorial file 12hr_topgenes.txt in the Query box.
 make sure that your list contains only official gene symbol (HUGO)
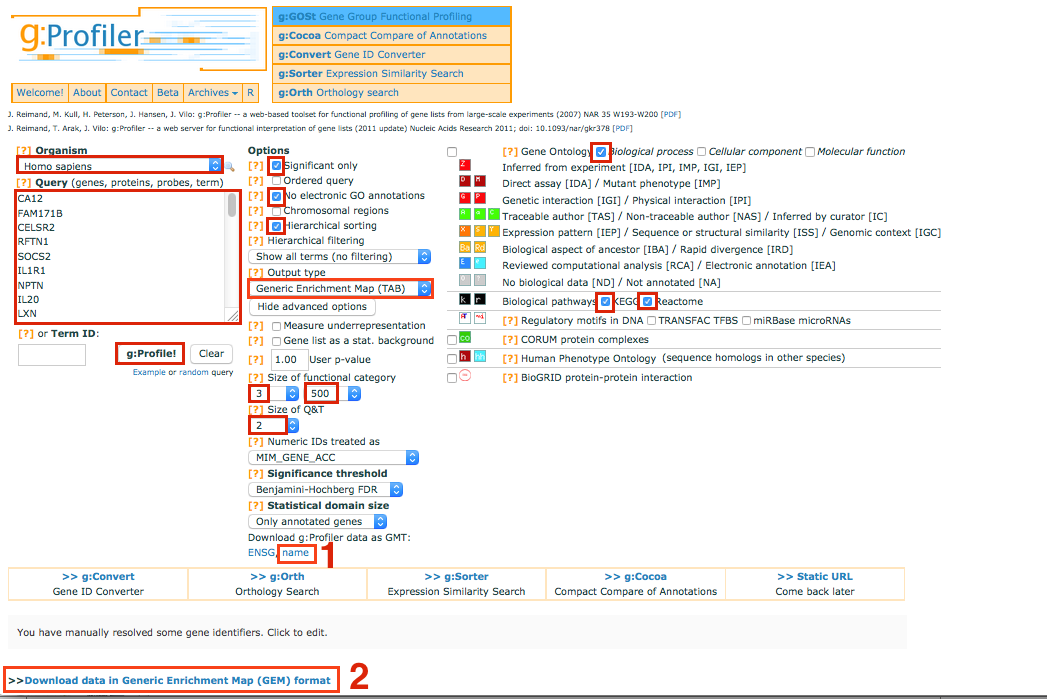
make sure that your list contains only official gene symbol (HUGO) In Options, check Significant only, No electronic GO annotations
Set the Output type to Generic EnrichmentMap
Show advanced options
Set Min and Max size of functional category to 3 and 500 respectively.
Select 2 for Size of Q&T
On the right panel, choose the Gene Ontology Biological process and Kegg and Reactome
Set Significance threshold to Benjamini-Hochberg FDR
Click on g:Profile! to run the analysis
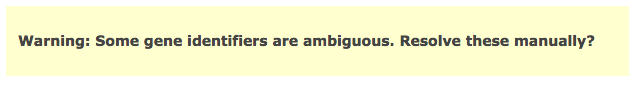
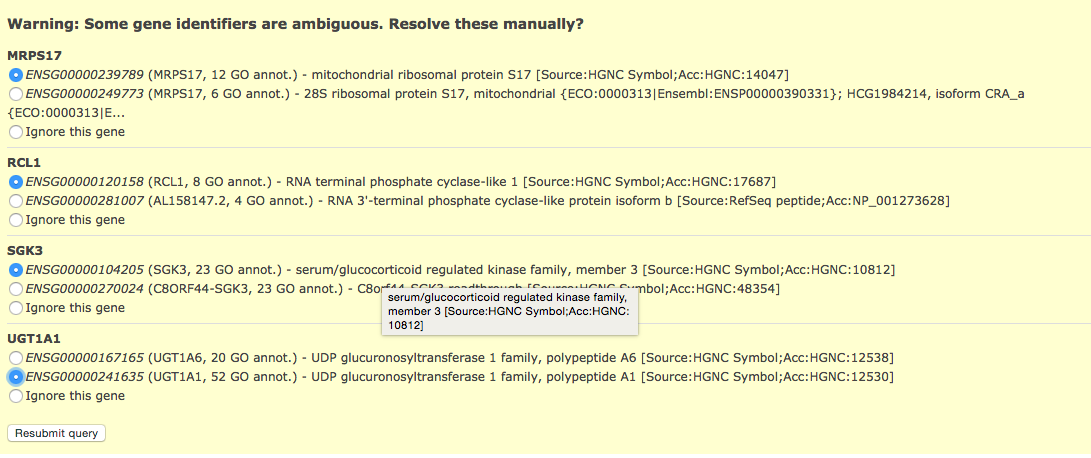
[Note] - if some of your identifiers in your query have multiple mappings in g:Profiler! by default they get excluded. If this happens you will see the following above the g:Profiler! results:
- Click on above link to manually map each gene to its correct annotation
click on Resubmit query to update your results with the specified mappings.
If the identifier discrepancy warning is ignored there might be differences between the number of genes g:Profiler attributes to a particular gene set and those associated with it in the Enrichment Map.
Download g:Profiler data as gmt: name Note: you will have to unzip the folder
Download the result file: Download data in Generic Enrichment Map (GEM) format
Note: repeat these steps for the 24hrs time-point and the file 24hr_topgenes.txt
Link to a step by step tutorial: gProfiler_step_by_step.pdf
Step 2: Generate Enrichment Map with g:Profiler Output
g:Profiler output files gProfiler_EM.zip
1. Open Cytoscape
2. In the menu bar, locate the App tab and then select --> EnrichmentMap --> Create Enrichment Map
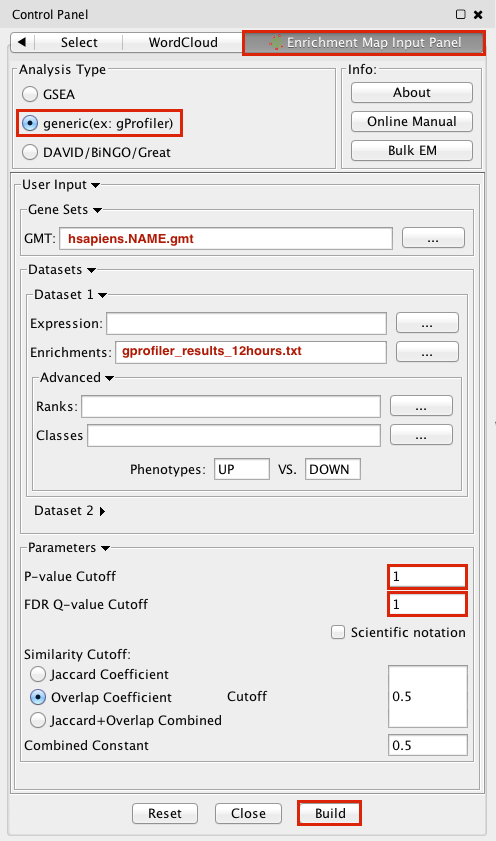
3. Make sure the Analysis Type is set to generic(ex:gProfiler)
4. Please select the following files by clicking on the respective (...) button and selecting the file in the Dialog:
GMT / hsapiens.NAME.gmt
Dataset 1 / Enrichments: gProfiler_results_12hr.txt
5. Tune Parameters
P-value cut-off 1
Q-value cut-off 1
Overlap Coefficient cut-off 0.5
6. Click on the Build radio button at the bottom of the panel to create the Enrichment Map
7. In the menu bar, Go to View, and activate Show Graphics Details
Step 3: Examining Results
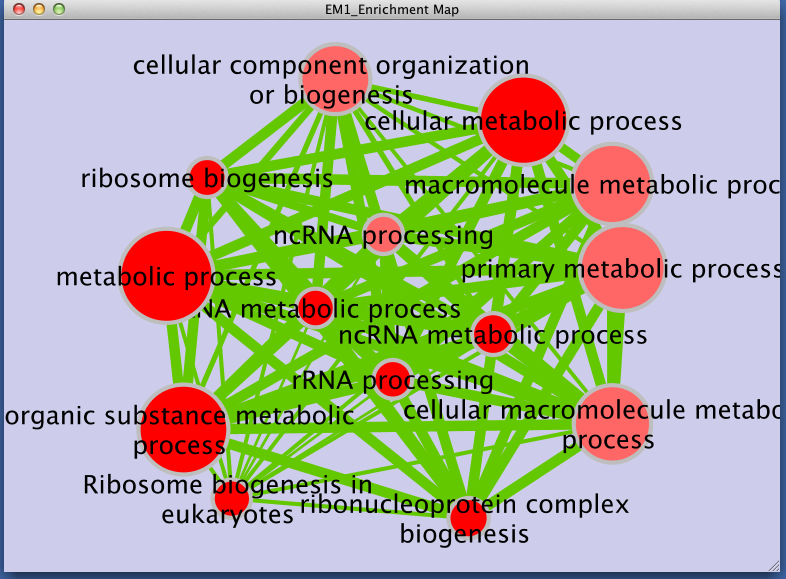
Legend:
- Node size corresponds to the number of genes in dataset 1 within the geneset
- Colour of the node corresponds to the significance of the geneset for dataset 1.
- Edge size corresponds to the number of genes that overlap between two connected genesets.