Examples of published papers that used GSEA and Enrichment Map
Example 1
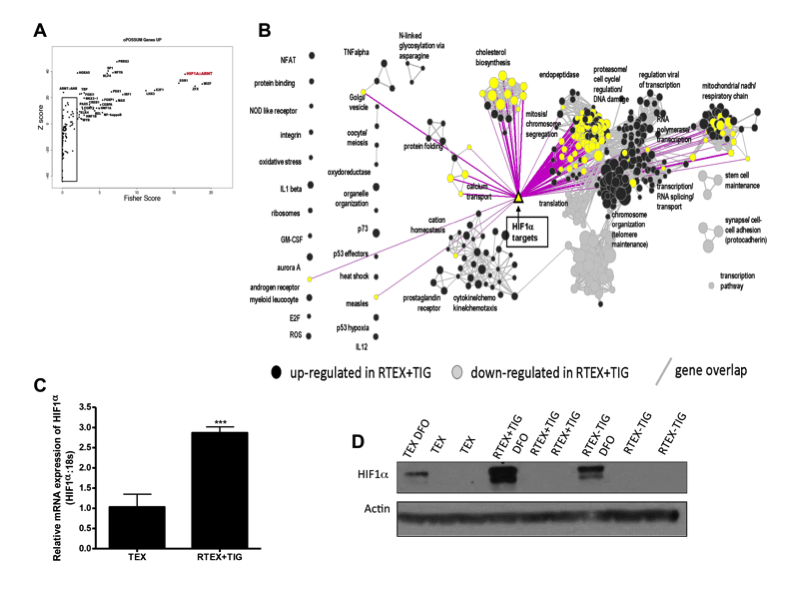
paper: Metabolic adaptation to chronic inhibition of mitochondrial protein synthesis in acutemyeloid leukemia cells. Jhas B, Sriskanthadevan S, Skrtic M, Sukhai MA, Voisin V, Jitkova Y, Gronda M, Hurren R, Laister RC, Bader GD, Minden MD, Schimmer AD.PLoS One. 2013;8(3):e58367. Epub 2013 Mar 8.
Example 2
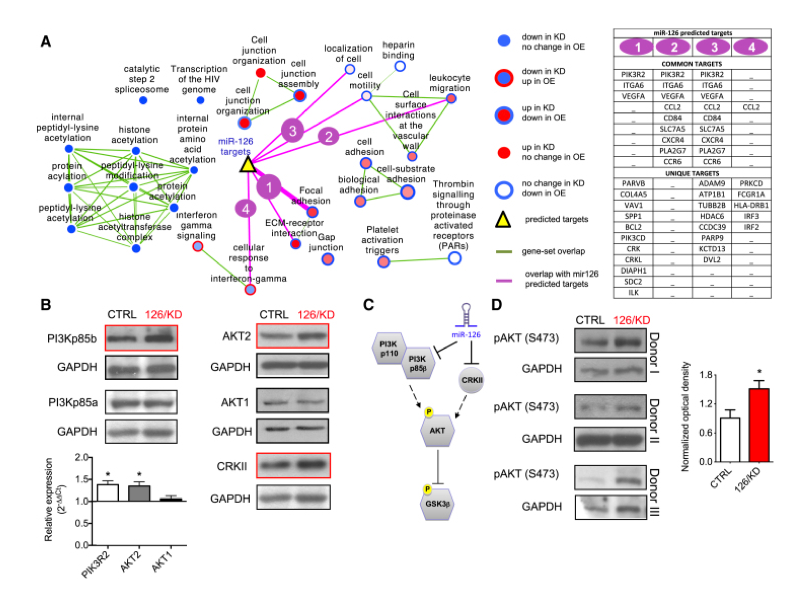
paper: Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Lechman ER, Gentner B, van Galen P, Giustacchini A, Saini M, Boccalatte FE, Hiramatsu H, Restuccia U, Bachi A, Voisin V, Bader GD, Dick JE, Naldini L.Cell Stem Cell. 2012 Dec 7;11(6):799-811.
Example 3
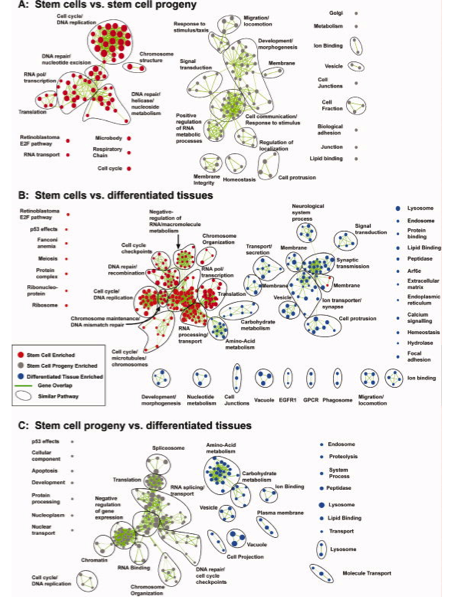
papaer: A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals.Labbé RM, Irimia M, Currie KW, Lin A, Zhu SJ, Brown DD, Ross EJ, Voisin V, Bader GD, Blencowe BJ, Pearson BJ. Stem Cells. 2012 Aug;30(8):1734-45.
Example 4
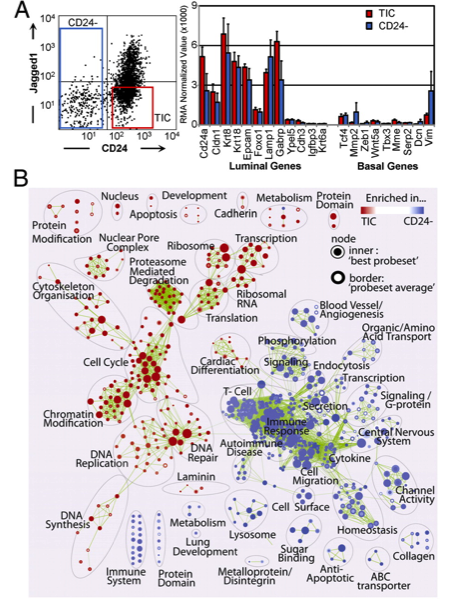
paper: Seventeen-gene signature from enriched Her2/Neu mammary tumor-initiating cells predicts clinical outcome for human HER2+:ERα- breast cancer. Liu JC, Voisin V, Bader GD, Deng T, Pusztai L, Symmans WF, Esteva FJ, Egan SE, Zacksenhaus E. Proc Natl Acad Sci U S A. 2012 Apr 10;109(15):5832-7. Epub 2012 Mar 28.