GSEA Gene Set Enrichment Analysis (www.broadinstitute.org/gsea )
Contents
-
GSEA Gene Set Enrichment Analysis (www.broadinstitute.org/gsea )
- Summary
- Introduction
- Method
- GSEA enrichment scores and statistics
- GSEA formats (.GCT, .CLS, .RNK, .GMT) :
- Tips on how to install GSEA locally and launch it from the command line:
- GSEA parameters
- GSEA tutorial
- Link to EnrichmentMap (Cytoscape) tutorials: how to create an enrichment map using GSEA results
- Link on how to use GSEA pre-ranked
- single sample GSEA (ssGSEA)
- References
- Answers to questions
- Troubleshooting
Summary
Gene Set Enrichment Analysis (GSEA) is a method for calculating gene-set enrichment.GSEA first ranks all genes in a data set, then calculates an enrichment score for each gene-set (pathway), which reflects how often members (genes) included in that gene-set (pathway) occur at the top or bottom of the ranked data set (for example, in expression data, in either the most highly expressed genes (top) or the most underexpressed genes(bottom).[adapted from http://www.broadinstitute.org/cancer/software/genepattern/modules/docs/GSEA/14]
Introduction
- GSEA has been first developped to interpret results from microarray experiments. One common approach to analyzing these data is to identify a limited number of the most interesting genes by picking a significance cutoff that will trim the list of interesting genes down to a handful of genes for further research. Gene Set Enrichment Analysis (GSEA) takes an alternative approach : it focuses on cumulative changes in expression of multiple genes as a group (belonging to a same gene-set/pahtway), which shifts the focus from individual genes to groups of genes. By looking at several genes at once, GSEA can identify pathways whose several genes each change a small amount, but in a coordinated way.
adapted from http://www.broadinstitute.org/cancer/software/genepattern/modules/docs/GSEA/14
Method
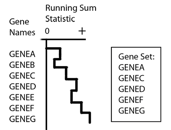
GSEA first ranks the genes based on a measure of each gene's differential expression with respect to the two phenotypes (for example, tumor versus normal using a t-test) or correlation with a continuous phenotype. Then the entire ranked list is used to assess how the genes of each gene set are distributed across the ranked list. To do this, GSEA walks down the ranked list of genes, increasing a running-sum statistic when a gene belongs to the set and decreasing it when the gene does not.
A simplified example is shown in the following figure.
The enrichment score (ES) is the maximum deviation from zero encountered during that walk. The ES reflects the degree to which the genes in a gene set are overrepresented at the top or bottom of the entire ranked list of genes. A set that is not enriched will have its genes spread more or less uniformly through the ranked list. An enriched set, on the other hand, will have a larger portion of its genes at one or the other end of the ranked list. The extent of enrichment is captured mathematically as the ES statistic.
Next, GSEA estimates the statistical significance of the ES by a permutation test. To do this, GSEA creates a version of the data set with phenotype labels randomly scrambled, produces the corresponding ranked list, and recomputes the ES of the gene set for this permuted data set. GSEA repeats this many times (1000 is the default) and produces an empirical null distribution of ES scores. Alternatively, permutations may be generated by creating “random” gene sets (genes randomly selected from those in the expression dataset) of equal size to the gene set under analysis.
The nominal p-value estimates the statistical significance of a single gene set's enrichment score, based on the permutation-generated null distribution. The nominal p-value is the probability under the null distribution of obtaining an ES value that is as strong or stronger than that observed for your experiment under the permutation-generated null distribution.
Typically, GSEA is run with a large number of gene sets. For example, the MSigDB collection and subcollections each contain hundreds to thousands of gene sets. This has implications when comparing enrichment results for the many sets:
The ES must be adjusted to account for differences in the gene set sizes and in correlations between gene sets and the expression data set. The resulting normalized enrichment scores (NES) allow you to compare the analysis results across gene sets.
The nominal p-values need to be corrected to adjust for multiple hypothesis testing. For a large number of sets (rule of thumb: more than 30), we recommend paying attention to the False Discovery Rate (FDR) q-values: consider a set significantly enriched if its NES has an FDR q-value below 0.25.
GSEA enrichment scores and statistics
GSEA_paper2005.pdf: equations and definition for statistics calculated by GSEA are described at the end of the paper as supplemental information.
- GSEA plot:
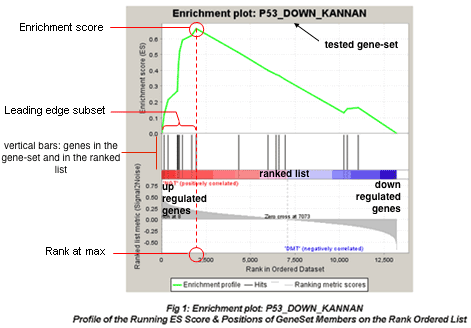
- which values to consider for interpreting results:
number of gene-set tested |
1 gene-set |
few gene-sets (3-10) |
a lot of gene-sets >3000 |
ES |
|
not recommended |
not recommended |
NES |
not informative |
|
|
nom p-value |
|
better to use FDR |
better to use FDR |
FDR |
not informative |
|
|
(*) FDR values are going to be pessimistic when using the BaderLab geneset files due to the high number of tested gene-sets and therefore the high p-value adjustment needed.
- ES (enrichment score): reflects the degree to which a gene-set is overrepresented at the top or bottom of a ranked list of genes.
- NES (normalized enrichment score): NES corrects for differences in ES between gene-sets due to differences in gene-set sizes. It enables to compare the scores of the different tested gene-sets with each other.
- NES = actual ES / mean of all ESs obtained from all random permutations for the single gene-set that is being tested
- nom p-value: The nominal p value estimates the statistical significance of the enrichment score for a single gene set. The p-value is calculated from the null distribution.
- Using gene-set permutation, the null distribution is created by generating, for each permutation, a random gene set the same size as your specified gene set by selecting that number of genes from all of the genes in your expression data set (or pre-ranked list), and then calculating the enrichment score for that randomly selected gene set. The distribution of those enrichment scores across all of the permutations constitutes the null distribution.
- FDR: corrects for multiple hypothesis testing and enable a more correct comparison of the different tested gene-sets with each other.
note: for a given gene-set S and observed NES, called NES*, FDR is [% of all NES (including permutations) >= NES*] / [% of all observed NES (=NES for all tested gene-sets) >= NES*]
- relationships between ES, pvalue , NES and FDR:
- pvalue is calculated from ES
- FDR is calculated from NES
- the higher the ES or NES and the lowest the FDR or pvalue
- NES above 1.4 will usually give significant results
GSEA formats (.GCT, .CLS, .RNK, .GMT) :
http://www.broadinstitute.org/cancer/software/gsea/wiki/index.php/Data_formats
http://www.broadinstitute.org/cancer/software/genepattern/modules/docs/GSEA/14
Tips on how to install GSEA locally and launch it from the command line:
- Download and Save the gsea2-2.0.14.jar file in your folder Documents
- open your console/terminal window
- Type the command for MAC:
- "java -Xmx2G -jar ~/Documents/gsea2-2.0.14.jar"
- Type the command for Windows:
- "cd Documents"
- "java -Xmx2G -jar gsea2-2.0.14.jar"
script that launches GSEA automatically on a MAC from the jar file (contains the gsea2-2.0.14.jar file): launch_gsea_mac_onlyversion.zip (download the zip file, unzip it, right click on the .command file and open it with Terminal)
script that launches GSEA automatically on a Windows machine from the jar file (contains the gsea2-2.0.14.jar file): launch_gsea_windows_onlyversion.zip (download the zip file, unzip and double click on the .bat file to launch GSEA).
GSEA parameters
GSEA tutorial
- step by step protocol using a .gct file as input file:
note: the .gct file used for this tutorial was generated using the CollapseDataset tool (menu bar --> Tools)
Link to EnrichmentMap (Cytoscape) tutorials: how to create an enrichment map using GSEA results
Guided GSEA tutorial to load the files into Enrichment Map
Guided GSEA/EM tutorial to create EM directly from GSEA interface for single analysis (direct link from GSEA Reports frame).
Guided GSEA/EM tutorial for multiple analyses to create an EM directly from GSEA interface for multiple analysis (from "Step in GSEA analysis frame)
Link on how to use GSEA pre-ranked
single sample GSEA (ssGSEA)
ssGSEA is available from the GenePattern website (http://www.broadinstitute.org/cancer/software/genepattern/)
- it takes a GCT file as input and it ranks each sample by the normalized in a descending order. ssGSEA performs a gene-set enrichment for each sample (=each column of the .GCT file) to see if genes at the top of the list are enriched in gene-sets in the gene-set database.
- the output is a new GCT file containing a NES (normalized enrichment score) for each sample and each tested gene-set. A heatmap done using the table of NES score helps for the interpretation of the results.
References
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102(43);15545-15550.
GSEA_paper2005.pdf: equations and definition for statistics calculated by GSEA are described at the end of the paper as supplemental information.
GSEA documentation: http://www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html
A more condensed document is available at http://www.broadinstitute.org/cancer/software/genepattern/modules/docs/GSEA/14
Answers to questions
- Question: how can we compare the NES (same gene-sets) between different datasets:
1) single sample GSEA case: e.g single sample GSEA was used on several patients and then a matrix of NES is created with the gene-sets as rows and patients as column and you want to find out gene-sets that are comparable between patients. A t-test with 1 group could be used to identify the gene-sets with comparable NES throughout samples --> t = mean/ standard error. The gene-sets will get a pvalue close to 0 only for gene-sets with comparable NES across patients (standard error is going to be small)
- 2) GSEA has been run using GSEA preranked option, you created a map using the 2 datasets and you see that the map is similar (e.g JAK2 responders versus partial responders) (high correlation throughout the gene-sets for the 2 datasets). You can do a K_S (Kolmogorov–Smirnov) test or a Wilcoxon rank sum test on the NES from the 2 datasets to see if "the 2 maps" are different or not.
Troubleshooting
- out of memory:
- running GSEA using the comprehensive Baderlab gene-set is using 1.8 GB of memory:
- Check that you have at least 2GB of free memory on your machine
- Check that you haved allocated enough memory to GSEA when launching GSEA
- using the javaGSEA Desktop Application: Launch with 2GB (for 64-bit Java only)
using the javaGSEA jar file: allocate 2GB of memory using this command: "java -Xmx2G –jar gsea2-2.0.14.jar"
- running GSEA using the comprehensive Baderlab gene-set is using 1.8 GB of memory:
- launching GSEA:
- GSEA works with java 6 , java 7 and java 8. However, for java 7 and 8, launching GSEA using the javaGSEA Desktop (java web start) may not work because on some security settings on your computer, in this case try to launch GSEA using the command line (javaGSEA jar file).
alternatively, you also can use GSEA from the GenePattern server (http://www.broadinstitute.org/cancer/software/genepattern/)