|
Size: 5810
Comment:
|
Size: 6115
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 9: | Line 9: |
| From this page you can learn more about the different datasets used to build the network shown in Figure 1b of the Primer about network visualization [Merico, D. Gfeller, D. & Bader, G.D., Nat. Biotech. x, x (2009), [[attachment:Primer.pdf|PDF]]] and download them if you want to try your own visualization or analyzing tools on them. | From this page you can learn more about the different datasets used to build the network shown in Figure 1b of the Primer about network visualization [Merico, D. Gfeller, D. & Bader, G.D., Nat. Biotech. x, x (2009), [[attachment:Primer.pdf|PDF]]] and download them if you want to try visualizing and analyzing them with your own tools. |
| Line 20: | Line 20: |
| Nodes represent proteins that are annotated as being located on the chromosome by the Gene Ontology project (for clarity, the suffix ‘p’ has been removed from yeast protein names). Node colors specify chromosomal location sub-categories: replication fork, red; nucleosome, green; kinetochore, blue; and other chromosome components, yellow. Edges represent protein-protein interactions that were manually extracted from publications by BioGRID database curators. Gene expression data of cells monitored during one round of the cell cycle are visually annotated on the network. Edge width encodes positive Pearson correlation between transcript profiles. Node size corresponds to the transcriptional amplitude, which is a measure of how much expression changes over the cell cycle. The network was visualized using the Cytoscape software. | Nodes represent proteins that are annotated as being located on the chromosome by the Gene Ontology project (for clarity, the suffix ‘p’ has been removed from yeast protein names). Node colors specify chromosomal location sub-categories: replication fork, red; nucleosome, green; kinetochore, blue; and other chromosome components, yellow. Edges represent protein-protein interactions that were manually extracted from publications by BioGRID database curators. Gene expression data of cells monitored during one round of the cell cycle are visually annotated on the network. Edge width encodes positive Pearson correlation between transcript profiles. Node size corresponds to the transcriptional amplitude, which is a measure of how much expression changes over the cell cycle. The network was visualized using the [[http://www.cytoscape.org|Cytoscape]] software. |
| Line 26: | Line 26: |
| * '''Protein-protein interactions''' [[attachment:protein-protein_interactions.sif|protein-protein_interaction.sif]]<<BR>>Proteins interactions are retrieved from the Biogrid database. Interactions between yeast proteins annotated as being located on the chromosome by the Gene Ontology (Cellular Component: chromosome and all its children) are listed. Biogrid interactions were filtered to keep only physical interactions describeded as "manually curated" (i.e. no data coming exclusively form automated literature mining have been included). Finally, we restricted ourselves to the proteins forming the largest component of the network. | * '''Protein-protein interactions''' [[attachment:protein-protein_interactions.sif|protein-protein_interaction.sif]]<<BR>>Proteins interactions are retrieved from the [[http://www.thebiogrid.org/|BioGRID]] database. Interactions between yeast proteins annotated as being located on the chromosome by the Gene Ontology (GO) (Cellular Component: chromosome and all its children) are listed. BioGRID interactions were filtered to keep only physical interactions described as "manually curated" (i.e. no data coming exclusively from automated literature mining have been included). Finally, we restricted ourselves to the proteins forming the largest connected component of the network. |
| Line 30: | Line 30: |
| * '''Yeast gene names''' [[attachment:yeast_gene_name.txt|yeast_gene_name.txt]]<<BR>> Systematic and standard name of the proteins. Splice variance were not included in this analysis. | * '''Yeast gene names''' [[attachment:yeast_gene_name.txt|yeast_gene_name.txt]]<<BR>> Systematic and standard name of the proteins. Splice variants were not included in this analysis. ('''are there any splice variants in this list? - these typically do not appear in yeast) |
| Line 32: | Line 32: |
| * '''Gene expression amplitude''' [[attachment:gene_expression_amplitude.txt|gene_expression_amplitude.txt]]<<BR>>Gene expression data come from [Spellman ''et al.'' Mol. Biol. Cell '''9''', 3273-3279 (1998)]. The gene expression amplitude was measured as the root mean square of the time-course expression values. If ''f(t)'' stands for the expression value of a given gene at time ''t'', the root mean square is computed as: | * '''Gene expression amplitude''' [[attachment:gene_expression_amplitude.txt|gene_expression_amplitude.txt]]<<BR>>Gene expression data come from [Spellman ''et al.'' Mol. Biol. Cell '''9''', 3273-3279 (1998)] ('''add link to pubmed'''). The gene expression amplitude was measured as the root mean square of the time-course expression values. If ''f(t)'' stands for the expression value of a given gene at time ''t'', the root mean square is computed as: ('''where is the formula?''') |
| Line 34: | Line 34: |
| * '''Gene expression correlation''' [[attachment:gene_expression_correlation.txt|gene_expression_correlation.txt]]<<BR>> For each pair of interacting proteins, we computed the Pearson correlation coefficient between the corresponding transcription profiles. Negative correlations were given a value of 0. The data in the attached file correspond to 1000 times the positive Pearson correlations. | * '''Gene expression correlation''' [[attachment:gene_expression_correlation.txt|gene_expression_correlation.txt]]<<BR>> For each pair of interacting proteins, we computed the Pearson correlation coefficient between the corresponding transcription profiles. Negative correlations were given a value of 0. The data in the attached file correspond to 1000 times the positive Pearson correlations. ('''why *1000?''') |
| Line 38: | Line 38: |
| 1. Download all the files listed above. | 1. Download all the files listed above and make sure you have Cytoscape installed and running smoothly. |
| Line 47: | Line 47: |
| 1. To add colors to the nodes, click on VizMapperTM button (you may need to click on the right arrow after the network button). In the Visual Mapping Browser double click "Node Color". Please select a value! appears on the right column: select "GO annotation". Please select a mapping!: choose "Discret Mapping". For each GO category, you can now choose the color you want. | 1. To add colors to the nodes, click on VizMapperTM button (you may need to click on the right arrow after the network button). In the Visual Mapping Browser double click "Node Color". Please select a value! appears on the right column: select "GO annotation". Please select a mapping!: choose "Discrete Mapping". For each GO category, you can now choose the color you want. |
still under construction...
Welcome to our wiki page: ''How to visually interpret biological data using networks''
From this page you can learn more about the different datasets used to build the network shown in Figure 1b of the Primer about network visualization [Merico, D. Gfeller, D. & Bader, G.D., Nat. Biotech. x, x (2009), PDF] and download them if you want to try visualizing and analyzing them with your own tools.
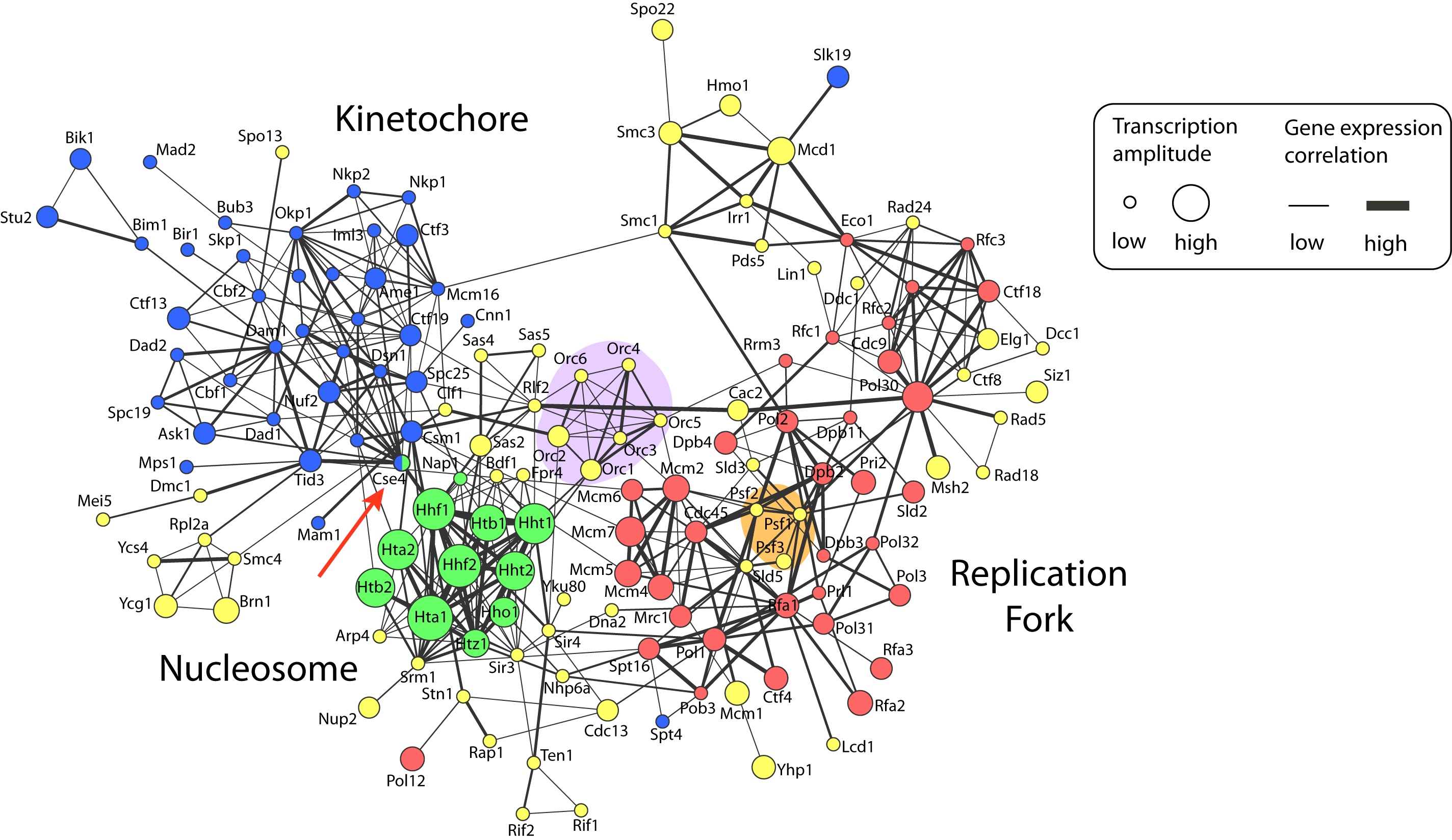
Figure 1: Network visualization of chromosome maintenance and duplication machinery in baker’s yeast, Saccharomyces cerevisiae (yeast_duplication_network.pdf).
Figure description
Nodes represent proteins that are annotated as being located on the chromosome by the Gene Ontology project (for clarity, the suffix ‘p’ has been removed from yeast protein names). Node colors specify chromosomal location sub-categories: replication fork, red; nucleosome, green; kinetochore, blue; and other chromosome components, yellow. Edges represent protein-protein interactions that were manually extracted from publications by BioGRID database curators. Gene expression data of cells monitored during one round of the cell cycle are visually annotated on the network. Edge width encodes positive Pearson correlation between transcript profiles. Node size corresponds to the transcriptional amplitude, which is a measure of how much expression changes over the cell cycle. The network was visualized using the Cytoscape software.
Datasets
The datasets described below can be freely downloaded. They have been formatted to be readily usable with Cytoscape software.
Protein-protein interactions protein-protein_interaction.sif
Proteins interactions are retrieved from the BioGRID database. Interactions between yeast proteins annotated as being located on the chromosome by the Gene Ontology (GO) (Cellular Component: chromosome and all its children) are listed. BioGRID interactions were filtered to keep only physical interactions described as "manually curated" (i.e. no data coming exclusively from automated literature mining have been included). Finally, we restricted ourselves to the proteins forming the largest connected component of the network.GO annotations GO_annotation.txt
Three sub-categories of the GO chromosome category were used to further specify the yeast proteins: replication fork (red), nucleosome (green) and kinetochore (blue). Cse4 is annotated to both the nucleosome and the kinetochore.Yeast gene names yeast_gene_name.txt
Systematic and standard name of the proteins. Splice variants were not included in this analysis. (are there any splice variants in this list? - these typically do not appear in yeast)Gene expression amplitude gene_expression_amplitude.txt
Gene expression data come from [Spellman et al. Mol. Biol. Cell 9, 3273-3279 (1998)] (add link to pubmed). The gene expression amplitude was measured as the root mean square of the time-course expression values. If f(t) stands for the expression value of a given gene at time t, the root mean square is computed as: (where is the formula?)Gene expression correlation gene_expression_correlation.txt
For each pair of interacting proteins, we computed the Pearson correlation coefficient between the corresponding transcription profiles. Negative correlations were given a value of 0. The data in the attached file correspond to 1000 times the positive Pearson correlations. (why *1000?)
Visualizing the network using Cytoscape
- Download all the files listed above and make sure you have Cytoscape installed and running smoothly.
- Open Cytoscape.
- Go to File/Import/network(multiple file types) and select "protein_protein_interaction.sif".
- Go to File/Import/Node Attributes and select "GO_annotation.txt".
- Go to File/Import/Node Attributes and select "yeast_gene_names.txt".
- Go to File/Import/Node Attributes and select "gene_expression_amplitude.txt".
- Go to File/Import/Edge Attributes and select "gene_expression_correlation.txt".
- To layout the network, go to Layout/Cytoscape Layouts/Force-directed Layouts/unweighted. You can try other layouts as well.
- Go to View/Show Graphics Details to see the original gene names.
- To add colors to the nodes, click on VizMapperTM button (you may need to click on the right arrow after the network button). In the Visual Mapping Browser double click "Node Color". Please select a value! appears on the right column: select "GO annotation". Please select a mapping!: choose "Discrete Mapping". For each GO category, you can now choose the color you want.
- To display the standard protein names as node labels, double click "Node Label". Select "Yeast gene names" and choose "Passthrough Mapping".
- To change the size of the nodes according to their gene expression amplitude, double click on "Node Size". Select "Gene expression amplitude". Choose "Continuous Mapping". Clicking on the chart that appears, you can now choose what node size you wish for the different amplitude values.
- To change the width of the edges according to the gene expression correlation, double click on Edge Size. Select "Gene expression correlation" and choose "Continuous Mapping". You can now choose what edge width you wish for the different correlation values.
Many other options to improve the layout or add other kind of visual attributes are available within Cytoscape. Have fun playing with them!
