|
Size: 38
Comment:
|
Size: 8401
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| Describe Software/HyperModules here. | <<TableOfContents(4)>> = HyperModules App = == Description == Hypermodules is a local graph search algorithm designed by Juri Reimand and Gary Bader. It was implemented in a command line version and as an app for [[http://www.cytoscape.org/ | Cytoscape 3.0]] as part of Google Summer of Code 2013. Given a gene/protein interaction network, a set of mutation data with associated patients, and a set of clinical patient data, the algorithm aims to find modules within the interaction network most correlated with a clinical outcome. In particular, survival times are analyzed using the log rank test for survival curve comparison, and fisher's exact test is used for discrete clinical variables. Only pSNV's are considered, and the algorithm can be applied to many clinical variables. For more info, please consult the original [[http://www.nature.com/msb/journal/v9/n1/full/msb201268.html| paper]]; the algorithm is described in the second half. == Command-Line Version == The lightweight command line version of the app is implemented in Java and compiled as an executable jar. To run, please ensure you have the latest version of Java installed on your machine. Then, navigate to the folder containing the jar file and run with the following command: java -jar [nameofjar.jar] [PATH_TO_NETWORK] [PATH_TO_MUTATION_DATA] [PATH_TO_CLINICAL_DATA] [SHUFFLE_NUMBER] [STATISTICAL_TEST] The shuffle number parameter should be between 100 and 5000, and the statistical test parameter is either "logrank" for survival data or "fisher" for data for a clinical variable. For example, using the attached example input files, assuming we have survival data and we want to do random shuffling of mutation associations 1000 times for accurate FDR discovery rate p-values, we run something that looks like this: java -jar HyperModulesCommandLine-0.0.1-SNAPSHOT-jar-with-dependencies.jar /Users/user/HyperModules/allinteractions.csv /Users/user/HyperModules/mutation_data.csv /Users/user/HyperModules/clinical_data.csv 1000 logrank. After the algorithm has finished running (NOTE: THIS MAY TAKE A LONG TIME, DEPENDING ON THE DENSITY OF THE TOPOLOGY OF THE INTERACTION NETWORK AND THE SIZE OF THE MUTATION DATA), there are only three basic options: enter 0 to export the results to a specified filepath, after providing the p-value cutoff you want to consider (to save all data, use a p-value cutoff of 1), enter 1 to print the results to screen (again, after entering a p-value cutoff), or enter 2 to exit the running program (all results data will be lost). * Download here: [[attachment:HMcmdline.tar | Download App]] == Cytoscape App Version == The full version of the app is implemented as a Cytoscape 3.X App (plugin). It will not work on earlier versions - to use, please sure you have the latest version of Cytoscape [[http://www.cytoscape.org/cy3.html | (Download here)]]! To install the app, * Go to the menu bar and click on '''Apps''' and then select '''App Manager''' * From here, either use ''' Search ''' to search for the App on the Cytoscape App Store, or choose ''' Install from File ''' to install the compiled version attached below * Click ''' Install from File ''', select the jar file in your local folder and select it. Once the app has been installed, go to ''' Apps ''' and select ''' HyperModules ''' from the dropdown, and then click ''' Open '''. The main panel should appear as a new tab in the left control panel of Cytoscape. To close the panel (and dispose of all results!), click ''' Close '''. If you click visualize, you can visualize a small portion of the interaction network currently selected in the Cytoscape viewer. To use it, enter a string into the text field of node names separated by commas and click execute (eg. "GENE1,GENE2,GENE3"). Here follows a brief overview of the options: {{attachment:hypermodules_panel.png|App User Panel|align="right"}} * In the ''' Select Network ''' panel, select the gene/protein interaction network that you want to run the algorithm on (if you already have one loaded in the cytoscape viewer). To add an interaction network to cytoscape, go to ''' File ''' -> ''' Import ''' -> ''' Network ''' -> ''' File... '''. * In the ''' Expand Option ''' panel, the default is to run the algorithm on all the seeds (every node in the network with at least one patient with that associated mutation). Select '''Expand from selected seeds ''' in order to only run the algorithm on the seeds that are currently selected (highlighted) in the Cytoscape network visualization window. * For ''' Analysis Type ''', select either ''' Survival ''' or ''' Discrete Variable ''' * For ''' Shuffle Number ''', enter a number between 0 and 5000. We recommend that you run the algorithm on a shuffle number of 1000. * In the ''' Load Mutation Data ''' panel, click on the button in order to load the mutation data file. Please follow the attached file to properly format your input file. There should be two columns - the first is all the gene/protein names, and the second is all the patients associated with (that have a mutation in) that gene. If there is no patient associated with that gene, it should be blank or it should say "no_sample". After you select the file, it should appear in the table that you can scroll through. Uncheck the ''' CSVHeaders''' option if your input file (CSV or TSV) doesn't have headers. ".maf" files are also acceptable input. * In the ''' Load Clinical Data ''' panel, click the button to load either clinical survival data (if you are running the log-rank test) or clinical variable data (if you are running fisher's exact test). After the file is loaded, it should appear in the scrollable table. By default, the first three columns of the CSV or TSV file will be used as the data. * For Survival Analysis, the first column should have all your patients, the second column should indicate their vital status (DECEASED/ALIVE, Y/N, or 1/0), and the third column should be the days to the last followup of the patient. * For Discrete, the first column should have all the patients, and the second column should have the patient's status with regards to the clinical variable of note. NOTE: if there are more than two kinds of values here, it may take a long time (fisher's exact test uses 2x2 tables by default). * To change the column for input data, select the appropriate column from the dropdown menu (which will display all of the columns in your input file). The changes will show up in the scrollable table. * Once everything is properly loaded, click ''' Run Algorithm '''. Again, this might take a long time depending on the topology of your interaction network and the size of the data. * When the algorithm is finished running, a panel should pop up in the right hand side in the Cytoscape results panel. Click the button with the triangle to get to the panel if you can't see the results panel. {{attachment:results_panel.png|Results Panel|align="right"}} * The table lists all the most correlated modules, subject to a p-value cutoff. Enter a new value in the text field and click '''Set P-Value Cutoff''' to change the cutoff. * Click ''' Export Results ''' to export the results into a csv file with all the data subject to the current cutoff (select a cutoff of 1 to save all data). * Click ''' Visualize Network ''' to view the network module of the row currently selected in the table. It will pop up as a new Cytoscape network. * Double click on ''' any row ''' in the table to view a graph of the Kaplan-Meier Survival Curves associated with the module (patients associated with the module versus all of the patients in your dataset). * Click ''' Discard Results ''' to exit the results panel. * Download here: COMING SOON == Sample Input Files == Please consult the following example files as a reference for the accepted format of the input files. Comma-separated values and tab-separated values are valid for input - avoid having any blank cells in your file. * Protein-Protein Interactions - [[attachment:allinteractions.csv | Interactions]] * Mutation Data - [[attachment:mutation_data.csv | Mutations]] * Patient Survival Data - [[attachment:clinical_data.csv| Survival]] * Patient Fisher Variable Data - [[attachment:vital_status.csv| Fisher]] |
HyperModules App
Description
Hypermodules is a local graph search algorithm designed by Juri Reimand and Gary Bader. It was implemented in a command line version and as an app for Cytoscape 3.0 as part of Google Summer of Code 2013. Given a gene/protein interaction network, a set of mutation data with associated patients, and a set of clinical patient data, the algorithm aims to find modules within the interaction network most correlated with a clinical outcome. In particular, survival times are analyzed using the log rank test for survival curve comparison, and fisher's exact test is used for discrete clinical variables. Only pSNV's are considered, and the algorithm can be applied to many clinical variables. For more info, please consult the original paper; the algorithm is described in the second half.
Command-Line Version
The lightweight command line version of the app is implemented in Java and compiled as an executable jar. To run, please ensure you have the latest version of Java installed on your machine. Then, navigate to the folder containing the jar file and run with the following command:
java -jar [nameofjar.jar] [PATH_TO_NETWORK] [PATH_TO_MUTATION_DATA] [PATH_TO_CLINICAL_DATA] [SHUFFLE_NUMBER] [STATISTICAL_TEST]
The shuffle number parameter should be between 100 and 5000, and the statistical test parameter is either "logrank" for survival data or "fisher" for data for a clinical variable. For example, using the attached example input files, assuming we have survival data and we want to do random shuffling of mutation associations 1000 times for accurate FDR discovery rate p-values, we run something that looks like this:
java -jar HyperModulesCommandLine-0.0.1-SNAPSHOT-jar-with-dependencies.jar /Users/user/HyperModules/allinteractions.csv /Users/user/HyperModules/mutation_data.csv /Users/user/HyperModules/clinical_data.csv 1000 logrank.
After the algorithm has finished running (NOTE: THIS MAY TAKE A LONG TIME, DEPENDING ON THE DENSITY OF THE TOPOLOGY OF THE INTERACTION NETWORK AND THE SIZE OF THE MUTATION DATA), there are only three basic options: enter 0 to export the results to a specified filepath, after providing the p-value cutoff you want to consider (to save all data, use a p-value cutoff of 1), enter 1 to print the results to screen (again, after entering a p-value cutoff), or enter 2 to exit the running program (all results data will be lost).
Download here: Download App
Cytoscape App Version
The full version of the app is implemented as a Cytoscape 3.X App (plugin). It will not work on earlier versions - to use, please sure you have the latest version of Cytoscape (Download here)!
To install the app,
Go to the menu bar and click on Apps and then select App Manager
From here, either use Search to search for the App on the Cytoscape App Store, or choose Install from File to install the compiled version attached below
Click Install from File , select the jar file in your local folder and select it.
Once the app has been installed, go to Apps and select HyperModules from the dropdown, and then click Open . The main panel should appear as a new tab in the left control panel of Cytoscape. To close the panel (and dispose of all results!), click Close . If you click visualize, you can visualize a small portion of the interaction network currently selected in the Cytoscape viewer. To use it, enter a string into the text field of node names separated by commas and click execute (eg. "GENE1,GENE2,GENE3"). Here follows a brief overview of the options:
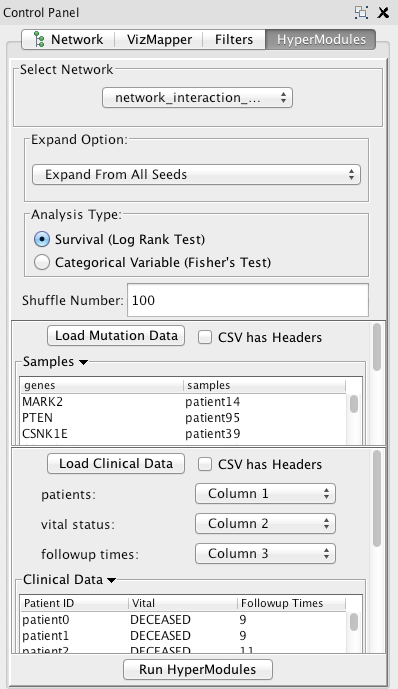
In the Select Network panel, select the gene/protein interaction network that you want to run the algorithm on (if you already have one loaded in the cytoscape viewer). To add an interaction network to cytoscape, go to File -> Import -> Network -> File... .
In the Expand Option panel, the default is to run the algorithm on all the seeds (every node in the network with at least one patient with that associated mutation). Select Expand from selected seeds in order to only run the algorithm on the seeds that are currently selected (highlighted) in the Cytoscape network visualization window.
For Analysis Type , select either Survival or Discrete Variable
For Shuffle Number , enter a number between 0 and 5000. We recommend that you run the algorithm on a shuffle number of 1000.
In the Load Mutation Data panel, click on the button in order to load the mutation data file. Please follow the attached file to properly format your input file. There should be two columns - the first is all the gene/protein names, and the second is all the patients associated with (that have a mutation in) that gene. If there is no patient associated with that gene, it should be blank or it should say "no_sample". After you select the file, it should appear in the table that you can scroll through. Uncheck the CSVHeaders option if your input file (CSV or TSV) doesn't have headers. ".maf" files are also acceptable input.
In the Load Clinical Data panel, click the button to load either clinical survival data (if you are running the log-rank test) or clinical variable data (if you are running fisher's exact test). After the file is loaded, it should appear in the scrollable table. By default, the first three columns of the CSV or TSV file will be used as the data.
- For Survival Analysis, the first column should have all your patients, the second column should indicate their vital status (DECEASED/ALIVE, Y/N, or 1/0), and the third column should be the days to the last followup of the patient.
- For Discrete, the first column should have all the patients, and the second column should have the patient's status with regards to the clinical variable of note. NOTE: if there are more than two kinds of values here, it may take a long time (fisher's exact test uses 2x2 tables by default).
- To change the column for input data, select the appropriate column from the dropdown menu (which will display all of the columns in your input file). The changes will show up in the scrollable table.
Once everything is properly loaded, click Run Algorithm . Again, this might take a long time depending on the topology of your interaction network and the size of the data.
- When the algorithm is finished running, a panel should pop up in the right hand side in the Cytoscape results panel. Click the button with the triangle to get to the panel if you can't see the results panel.
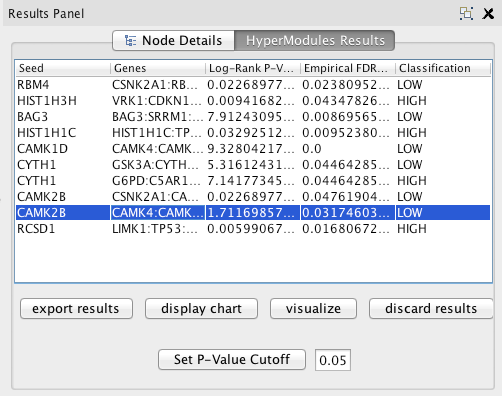
The table lists all the most correlated modules, subject to a p-value cutoff. Enter a new value in the text field and click Set P-Value Cutoff to change the cutoff.
Click Export Results to export the results into a csv file with all the data subject to the current cutoff (select a cutoff of 1 to save all data).
Click Visualize Network to view the network module of the row currently selected in the table. It will pop up as a new Cytoscape network.
Double click on any row in the table to view a graph of the Kaplan-Meier Survival Curves associated with the module (patients associated with the module versus all of the patients in your dataset).
Click Discard Results to exit the results panel.
- Download here: COMING SOON
Sample Input Files
Please consult the following example files as a reference for the accepted format of the input files. Comma-separated values and tab-separated values are valid for input - avoid having any blank cells in your file.
Protein-Protein Interactions - Interactions
Mutation Data - Mutations
Patient Survival Data - Survival
Patient Fisher Variable Data - Fisher
