Google Summer of Code 2010: Semantic Network Summary
Short Description (from GenMAPP wiki)
Goal
Develop a visual summary of a set of node attributes
Description
When biological networks are investigated, it is common to look for clusters, i.e. sets of nodes that are highly inter-connected. To figure out the "biological meaning" of a cluster, the user has to sift through the long textual annotations that are associated to biological entities. We are interested in producing a graphical summary of such annotations. Word frequency in annotations is a good starting point. This can be visualized as a "tag cloud". In addition, the word layout can reflect similarity relations among words (e.g. co-occurrence in the same annotations).
This functionality can be applied to several networks outside biology, whenever nodes are associated to verbose textual annotation. An example is professional social networks (e.g. linked-in), where individuals are "annotated" by a short CV.
Language and Skills
Java, basic statistics
Longer Description
Problem Description
Biological networks can be visualized and analyzed using Cytoscape. Since biological networks have a large number of nodes (a network consisting of all the proteins in a cell can have up to 20k nodes), one of the common approaches to summarize a network is by clustering, i.e. identifying groups of highly inter-connected nodes. Clusters can be identified algorithmically, or hand-picked by experts (with the help of the network layout).
Once clusters have been identified, however, it not trivial to summarize their meaning. Bio-entities typically have rich semantics, which are encoded by long string attributes.
The purpose of the Semantic Network Summary module will be to generate more concise summaries.
Input/Output Description
This functionality should be implemented as a cytoscape plugin.
The plugin will receive as input (S, A), a set of nodes S = {s1, s2, ..., sn} together with their set of attributes A = {a1, a2, ..., an}.
The user can choose one or more of the attributes to be used to summarize the set of nodes (for example the user might want to use both name and description of each node to extract a summary)
For every input (S, A), a graphical or visual summary of the attributes will have to be generated and displayed within the cytoscape panel.
Attributes A can be any type of attribute associated with a cytoscape network(i.e. String, Int, Double, boolean list...)
Input Examples
An example network the plugin should work on can be found in the attached Cytoscape session file:
Other attribute examples:
Available Solutions: Word Frequency
A first simple solution we have implemented:
- break down {a1, a2, ..., an} into single words
- count word frequencies
- use coefficients based on information theory, or a statistical test p-value
This simple idea can be improved by:
- removing common-place words (e.g. "of", "by", etc...)
- dividing the word frequencies in A by the word frequencies in the full network (i.e. all nodes)
Wordle is a cool graphical representation based on word frequency.
Here's how we used wordle to generate a semantic summary in one of our papers (string attributes are displayed as node labels).
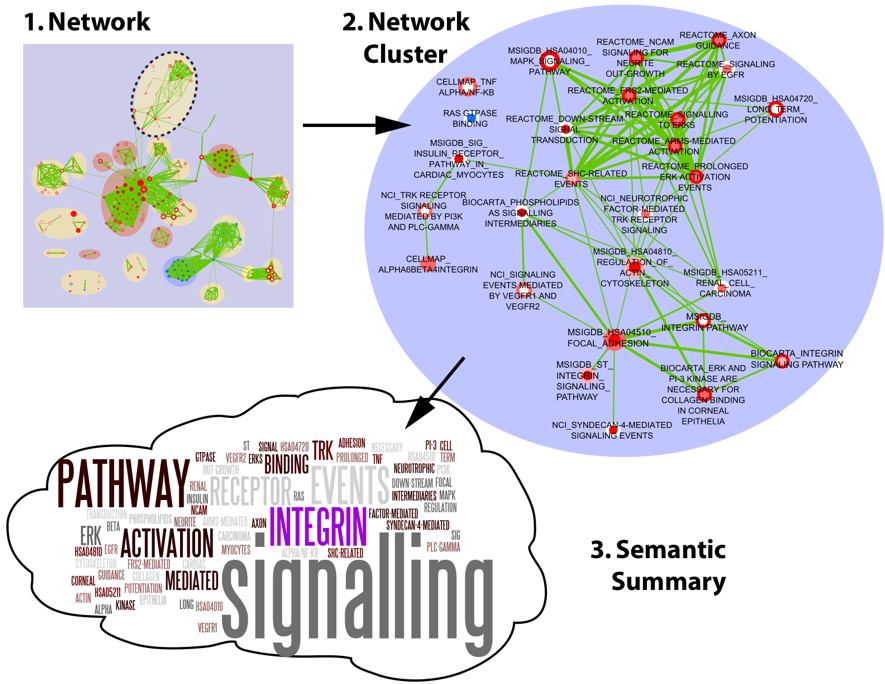
Ref: Isserlin R, Merico D, Alikhani-Koupaei R, Gramolini A, Bader GD, Emili A. Pathway analysis of dilated cardiomyopathy using global proteomic profiling and enrichment maps. Proteomics. 2010 Feb 1. PMID: 20127684
Going Beyond Simple Solutions
We would like applicants to be creative, and come up with new or different ways to improve the frequency-based semantic summary.
We think taking into account relations between words would be very useful to make the semantic summary richer and more informative. In fact, breaking down description into words can make it harder to grasp the original meaning of string attributes.
Environment: Cytoscape
- We want the semantic network summary to be implemented as a Cytoscape plugin
- Cytoscape plug-ins are coded in Java using the Cytoscape API
About
This project was started by
We are part of Gary Bader's lab at University of Toronto - CCBR (Toronto, ON Canada). Our lab is strongly engaged in biological network research. Feel free to have a look at our home page for more details on the lab research areas, and at our home-pages for our own research interests. Here is also a Cytoscape plugin we have recently developed.