|
Size: 854
Comment:
|
Size: 1573
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 13: | Line 13: |
| == openPIP Featurs == | == openPIP Features == * Data deposition is the PSI-MI-TAB format, the PPIs standard format. * Hasel-free customization, change the look and contents for the admin tool, no coding required. * Comprehensive search options. * Search results visualization using [[http://js.cytoscape.org/|Cytoscap.JS]] * Downloading search results in five different formats including TSV, PSI-MI-TAB, PNG and JPG. * Automated post/delete news and announcements. <<BR>>{{attachment:openPIP-HomePage.png|openPIP homepage|align="center"}}<<BR>> '''openPIP homepage:''' The homepage contents are editable through the admin tool. <<BR>>This includes the main text, footer, announcments and news, color theme, database name, database short name and logo colors. |
| Line 15: | Line 27: |
| CategoryBuildings |
openPIP: The Open Source Protein Interactions Platform

Brief Description
The Open Source Protein Interactions Platform (openPIP) is an open source software package for hosting protein-protein interaction (PPI) data based on the PSI-MI-TAB format. openPIP provides PPI database and a customizable web interface that enables searching and visualizing the interactions as well as downloading the search results in different formats. The whole system is a ready-to-use virtual machine that can be imported to VMware.
A live version of openPIP is available at this link.
openPIP Features
- Data deposition is the PSI-MI-TAB format, the PPIs standard format.
- Hasel-free customization, change the look and contents for the admin tool, no coding required.
- Comprehensive search options.
Search results visualization using Cytoscap.JS
- Downloading search results in five different formats including TSV, PSI-MI-TAB, PNG and JPG.
- Automated post/delete news and announcements.
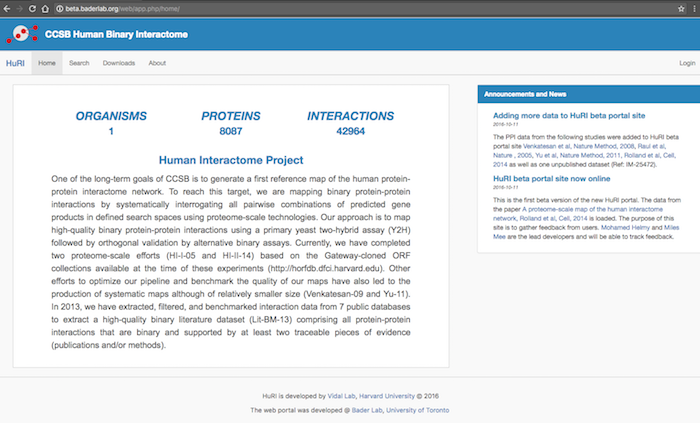
openPIP homepage: The homepage contents are editable through the admin tool.
This includes the main text, footer, announcments and news, color theme, database name, database short name and logo colors.
