|
Size: 1573
Comment:
|
Size: 1754
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 4: | Line 4: |
| = openPIP: The Open Source Protein Interactions Platform = | = openPIP: The Open Source Protein Interaction Platform = |
| Line 9: | Line 9: |
| The Open Source Protein Interactions Platform (openPIP) is an open source software package for hosting protein-protein interaction (PPI) data based on the [[https://code.google.com/archive/p/psimi/wikis/PsimiTab27Format.wiki|PSI-MI-TAB format]]. openPIP provides PPI database and a customizable web interface that enables searching and visualizing the interactions as well as downloading the search results in different formats. The whole system is a ready-to-use virtual machine that can be imported to VMware. | The Open Source Protein Interaction Platform (openPIP) is an open source software package for hosting protein-protein interaction (PPI) data based on the [[https://code.google.com/archive/p/psimi/wikis/PsimiTab27Format.wiki|PSI-MI-TAB format]]. openPIP provides PPI database and a customizable web interface that enables searching and visualizing the interactions as well as downloading the search results in different formats. The whole system is a ready-to-use virtual machine that can be imported to VMware. |
| Line 23: | Line 23: |
| '''openPIP homepage:''' The homepage contents are editable through the admin tool. <<BR>>This includes the main text, footer, announcments and news, color theme, database name, database short name and logo colors. | '''openPIP Homepage:''' The homepage contents are editable through the admin tool. <<BR>>This includes the main text, footer, announcments and news, color theme, database name, database short name and logo colors. <<BR>>{{attachment:openPIP-AdminSettings.png|openPIP Settings|align="center"}}<<BR>> '''openPIP Global Settings:''' The global settings anables costomization of the website look. |
openPIP: The Open Source Protein Interaction Platform

Brief Description
The Open Source Protein Interaction Platform (openPIP) is an open source software package for hosting protein-protein interaction (PPI) data based on the PSI-MI-TAB format. openPIP provides PPI database and a customizable web interface that enables searching and visualizing the interactions as well as downloading the search results in different formats. The whole system is a ready-to-use virtual machine that can be imported to VMware.
A live version of openPIP is available at this link.
openPIP Features
- Data deposition is the PSI-MI-TAB format, the PPIs standard format.
- Hasel-free customization, change the look and contents for the admin tool, no coding required.
- Comprehensive search options.
Search results visualization using Cytoscap.JS
- Downloading search results in five different formats including TSV, PSI-MI-TAB, PNG and JPG.
- Automated post/delete news and announcements.
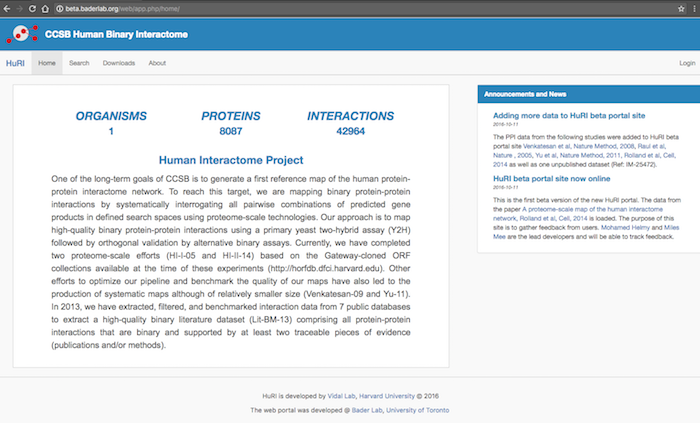
openPIP Homepage: The homepage contents are editable through the admin tool.
This includes the main text, footer, announcments and news, color theme, database name, database short name and logo colors.
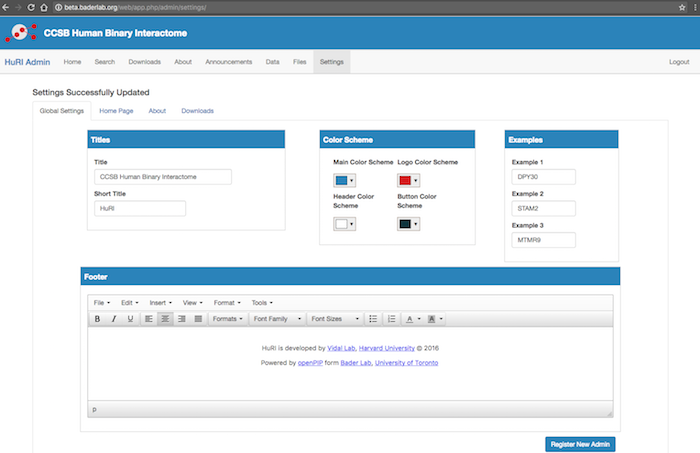
openPIP Global Settings: The global settings anables costomization of the website look.
