|
Size: 6739
Comment:
|
Size: 6437
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| #acl All:read | |
| Line 3: | Line 4: |
| LOLA is currently in beta-testing. Version 1.0-beta is now available for download. | LOLA is currently in beta-testing. Version 1.36 is now available for download. |
| Line 5: | Line 6: |
| [[TableOfContents()]] | <<TableOfContents>> |
| Line 7: | Line 8: |
| === Latest Download === | ---- == Downloads == === Latest Release === ''LOLA Version 1.36 (April 4, 2012)'' Build (verified to run with jre 1.6.0_22): [[attachment:LOLA-1.36.zip||&do=get]] |
| Line 9: | Line 14: |
| LOLA Version 1.0 Beta: [attachment:lola-1.0-beta.tar Download] | === Current Development Release === ''LOLA Version 1.36 (April 4, 2012)'' Source + dependencies (verified to run with jre 1.6.0_22): [[attachment:LOLA-1.36-src.zip||&do=get]] |
| Line 11: | Line 18: |
| === Requirements === | === Earlier Stable Releases === ''LOLA Version 1.35 Alpha (June 5, 2009)''<<BR>> Build: [[attachment:LOLA-1.35-alpha.zip||&do=get]] (verified to run with jre 1.6.0_22) |
| Line 13: | Line 21: |
| Java Runtime Environment (JRE) 1.5 or later is required to run LOLA. All other dependencies are included in the download. | ''LOLA Version 1.3 Beta (September 29, 2008)''<<BR>> Build: [[attachment:LOLA-1.3-beta.tgz]] <<BR>> Source: [[attachment:LOLA-1.3-beta-src.tgz||&do=get]] |
| Line 15: | Line 23: |
| === Installing and Running === | ''LOLA Version 1.2 Beta (May 12, 2008)''<<BR>> Build: [[attachment:LOLA-1.2-beta.tgz]] <<BR>> Source: [[attachment:LOLA-1.2-beta-src.tgz]] ''LOLA Version 1.1 Beta (August 22, 2007)''<<BR>> Build: [[attachment:LOLA-1.1-beta.tgz]] <<BR>> Source: [[attachment:LOLA-1.1-beta-src.tgz]] === Release Notes === '''Version 1.36:''' * Position weight matrices in PWM file format can now be loaded (please see file formats below). * Users can choose to draw logos with or without axis labels via Logo Options -> Basic. '''Version 1.35 alpha:''' * Java Runtime Environment (JRE) 1.6 is required to run LOLA (verified to run with jre 1.6.0_22). All other dependencies are included in the download. * For each occurrence of the symbol 'X' (which represents an ambiguous amino acid) at a position in the profile, contribute an equal 1/20 weight to each residue in the corresponding weight distribution. Previously, the weight contributed to each residue was 1.0, causing inflation of "background" signal and hence suppression of significant features at the given position. '''Version 1.3 beta:''' * Progress bars now appear for long-running tasks such as loading peptide file and generating logos and logo trees * Notable bug fixes: * Selected profiles now correctly closed with the "Close Selected" button. * Regenerating a logo tree after closing some profiles now correctly excludes the closed profiles. '''Version 1.2 beta:''' * Logo trees can be generated for all types of domains - not only terminal binding * New advanced options including amino acid coloring styles, amino acid grouping, and logo tree leaf ordering '''Version 1.1 beta:''' * New ''Logo Tree'' feature - basic functionality for PDZ or other terminal binding motifs not requiring profile alignment ---- == Installing and Running == '''Requirements:''' Java Runtime Environment (JRE) 1.5 or later is required to run LOLA. All other dependencies are included in the download. |
| Line 22: | Line 61: |
| * You can open a single peptide file, or a project file linked to multiple peptide files. | LOLA accepts one or more [[Software/BRAIN/PeptideFile|peptide file]] as shown in the example below. A peptide file describes a protein containing a specific domain, and provides known peptide ligands of this domain obtained by an experimental technique. You can open a single peptide file, or multiple peptide files grouped into a [[Software/BRAIN/PeptideFile#ProjectFiles|project file]]. |
| Line 24: | Line 63: |
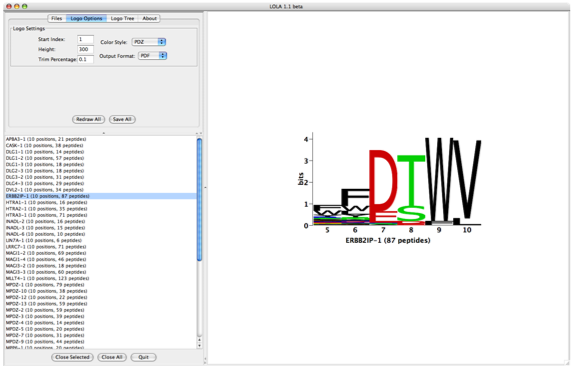
| Here's a view of LOLA after opening a PDZ domain project file: | LOLA also accepts one or more [[Software/LOLA/PWMFile|pwm file]] as shown in the example below. A pwm file describes a position weight matrix (rows are amino acids, columns are ligand positions) where each cell in the matrix represents the frequency of observing a specific amino acid at a given ligand position. You can open a single pwm file, or multiple pwm files grouped into a [[Software/LOLA/PWMFile#ProjectFiles|project file]]. |
| Line 26: | Line 65: |
| attachment:lolaScreenShot.png | Here's a view of LOLA after opening a PDZ domain project file, and after generating a logo tree: |
| Line 28: | Line 67: |
| === Input Format === LOLA accepts one or more ''peptide file'' as shown in the example below. A peptide file describes a protein containing a specific domain, and provides known peptide ligands of this domain obtained by an experimental technique. |
{{attachment:lolaScreenShot-Small.png}} {{attachment:lolaScreenShotLogoTree.png}} |
| Line 31: | Line 69: |
| The peptide file consists of a '''Header Section''' that describes the protein and domain sequence, and a '''Peptide Section''' that lists and describes the peptide ligands. | === Changing memory allocations on Windows, Mac, and Linux machines === There are a number of ways to change LOLA's memory allocation, depending on your preferred method of opening the application. |
| Line 33: | Line 72: |
| Example: {{{ Gene Name DLG1 Accession Refseq:NP_004078 Organism Homo Sapiens (Human) NCBITaxonomyID 9606 Domain Number 3 Domain Type PDZ Interpro ID IPR001478 Technique Phage Display High Valency Domain sequence KVVLHRGSTGLGFNIVGGEDGEGIFISFILAGGPADLSGELRKGDRIISVNSVDLRAASHEQAAAALKNAGQAVTIVA Domain Range 466-525 Comment PeptideName Peptide CloneFrequency QuantData ExternalIdentifier 1 XLHFWRESSV 66 2 XXRLWKQTSL 3 3 ILKIWRETSL 3 4 KRTIWRETSL 2 A KNLRSNSMLG 2 6 HLKFWRSTRV 2 7 AHSKWRSTSV 2 8 XXXHRRETTV 1 9 VISRWRQTSL 1 10 TTWLGRQTRV 1 11 SRSSYRETSV 1 12 XXXSRRETSV 1 13 RLFRYRETSL 1 B PIRKRWTMTL 1 15 XXXNHRETSV 1 16 KIVRWKNTSV 1 17 KHRTWYETSV 1 18 XXXXFKQTSV 1 19 ARPKWRTTRV 1 20 ALPRRRETSV 1 }}} |
==== Option A: Command line startup (note: this does not permanently change LOLA's default 512M setting) ==== If you are opening LOLA from the command-line using the command: |
| Line 69: | Line 75: |
| ==== Header Section ==== | * java –Xmx512M –jar lola-VERSION.jar |
| Line 71: | Line 77: |
| Describes the protein, domain, and experiment. Required fields are indicated with a '''*'''. | then you can increase the value of –Xmx to the desired amount of memory. For example: |
| Line 73: | Line 79: |
| '''NOTE:''' This section is in a 2 column format. Field names must be separated from their values with a single TAB character. Multiple TABs or spaces are not accepted. | * java –Xmx800M –jar lola-VERSION.jar |
| Line 75: | Line 81: |
| '''Gene Name:*''' An identifier that represents the gene or protein sequence. Not required to be unique. | ==== Option B: Using lola.bat (Windows systems) ==== 1. Open the file lola.bat in a text editor (eg. right-click and select Open With Notepad). 1. Increase the value of the –Xmx tag (found in the last line of the file), as per Option A. Do not modify other parts of the file. 1. Save and close the file. 1. Open LOLA by double-clicking on lola.bat. |
| Line 77: | Line 87: |
| '''Accession:*''' A space-separated list of database accession identifiers for the protein or corresponding gene. | ==== Option C: Using lola.sh (UNIX, Linux, and Mac OS X systems) ==== 1. Open the file lola.sh in a text editor (eg. right-click and select Open With TextEdit). 1. Increase the value of the –Xmx tag (found in the last line of the file), as per Option A. Do not modify other parts of the file. 1. Save and close the file. 1. Open LOLA by running lola.sh from the command-line. |
| Line 79: | Line 93: |
| '''Organism:''' Description of taxon of the protein. | ---- == References == LOLA has been referenced in the following publications: |
| Line 81: | Line 97: |
| '''NCBITaxonomyID*:''' Taxon identifier from NCBI's Taxonomy repository. | Tonikian, R. |
| Line 83: | Line 99: |
| '''Domain Number*:''' A number that represents the position of the domain sequence within the protein. For proteins containing multiple instances of the domain, this number helps distinguish the position of these instances. Set to "0" if instance information is not known. | ''et al.''(2008) A Specificity Map for the PDZ Domain Family. |
| Line 85: | Line 101: |
| '''Domain Type:*''' The formal name of the domain, e.g. WW, PDZ, SH3. | ''PLOS Biology'', 6, 9: 2043-2059. |
| Line 87: | Line 103: |
| '''Interpro ID:''' The Interpro database identifier for the domain. | http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0060239 |
| Line 89: | Line 105: |
| '''Technique:''' The experimental method used to identify potential ligands of the protein. '''Domain Sequence:*''' The amino-acid sequence of the domain region. '''Domain Range:''' The amino-acid position range for the domain region within the protein. '''Comment:''' Notes, additional information, personal comments pertaining to this file. ==== Peptide Section ==== Describes the experimentally determined peptide ligands. The peptide sequences must be in '''multiple alignment format'''. The sequences should contain '''no gaps''', and should be padded with the '''X''' symbol on both sides, where required, such that all sequences have identical length. '''NOTE:''' This section is in a 5-column format. Column headers and values must be separated with a single TAB character. Multiple TABs or spaces are not accepted. Required fields are indicated with a '''*'''. '''PeptideName:*''' A ''unique'' numerical symbol assigned to each peptide ligand. To omit a peptide, set to a non-numeric value (e.g. "A"). '''Values in this column must be unique.''' '''Peptide:*''' The peptide ligand sequence. '''CloneFrequency:''' Applies only to phage display data: the observed frequency of the peptide in the cloning step. '''QuantData:''' A number that relatively or absolutely quantifies the protein-ligand interaction. E.g. The optical density (OD) from a protein chip experiment. '''ExternalIdentifier:''' A database identifier for the peptide. === Using Project Files === To open several peptide files at once, simply link them all in a single '''project file'''. A project file is a text file containing the absolute paths of multiple peptide files. Opening the project file in LOLA will open each of the underlying peptide files in a single step, allowing logos to be constructed for multiple profiles. Example: {{{ #ProjectFile /Users/moyez/research/ppi/profiles/PDZ/Human/SidhuPhage/APBA3-1.pep.txt /Users/moyez/research/ppi/profiles/PDZ/Human/SidhuPhage/CASK-1.pep.txt /Users/moyez/research/ppi/profiles/PDZ/Human/SidhuPhage/DLG1-1.pep.txt /Users/moyez/research/ppi/profiles/PDZ/Human/SidhuPhage/DLG1-2.pep.txt /Users/moyez/research/ppi/profiles/PDZ/Human/SidhuPhage/DLG1-3.pep.txt /Users/moyez/research/ppi/profiles/PDZ/Human/SidhuPhage/DLG2-3.pep.txt /Users/moyez/research/ppi/profiles/PDZ/Human/SidhuPhage/DLG3-2.pep.txt /Users/moyez/research/ppi/profiles/PDZ/Human/SidhuPhage/DLG4-3.pep.txt /Users/moyez/research/ppi/profiles/PDZ/Human/SidhuPhage/DVL2-1.pep.txt /Users/moyez/research/ppi/profiles/PDZ/Human/SidhuPhage/ERBB2IP-1-hi.pep.txt }}} '''NOTE:''' The first line of the project file '''must''' contain the text "#ProjectFile". === Future Developments === * Generate a "logo tree" by hiearchically clustering logos |
---- == Future Developments == * Enable logo and logo tree parameters to be saved in a project file for later use * Support customized amino acid grouping schemes |
| Line 144: | Line 113: |
| === Contact === If you have any questions or feedback, please email Moyez Dharsee at mdharsee@infochromics.com. |
---- == Contact == If you have any questions or feedback, please email Moyez Dharsee at mdharsee@infochromics.com . |
LOLA (LOgos Look Amazing) is a tool for generating sequence logos using Position Weight Matrix based protein profiles. LOLA allows you to generate custom sequence logos by setting parameters such as logo height, trim percentage, and residue colour scheme. You can then save logos in various formats including PDF, PNG, and JPEG.
LOLA is currently in beta-testing. Version 1.36 is now available for download.
Downloads
Latest Release
LOLA Version 1.36 (April 4, 2012) Build (verified to run with jre 1.6.0_22): LOLA-1.36.zip
Current Development Release
LOLA Version 1.36 (April 4, 2012) Source + dependencies (verified to run with jre 1.6.0_22): LOLA-1.36-src.zip
Earlier Stable Releases
LOLA Version 1.35 Alpha (June 5, 2009)
Build: LOLA-1.35-alpha.zip (verified to run with jre 1.6.0_22)
LOLA Version 1.3 Beta (September 29, 2008)
Build: LOLA-1.3-beta.tgz
Source: LOLA-1.3-beta-src.tgz
LOLA Version 1.2 Beta (May 12, 2008)
Build: LOLA-1.2-beta.tgz
Source: LOLA-1.2-beta-src.tgz
LOLA Version 1.1 Beta (August 22, 2007)
Build: LOLA-1.1-beta.tgz
Source: LOLA-1.1-beta-src.tgz
Release Notes
Version 1.36:
- Position weight matrices in PWM file format can now be loaded (please see file formats below).
Users can choose to draw logos with or without axis labels via Logo Options -> Basic.
Version 1.35 alpha:
- Java Runtime Environment (JRE) 1.6 is required to run LOLA (verified to run with jre 1.6.0_22). All other dependencies are included in the download.
- For each occurrence of the symbol 'X' (which represents an ambiguous amino acid) at a position in the profile, contribute an equal 1/20 weight to each residue in the corresponding weight distribution. Previously, the weight contributed to each residue was 1.0, causing inflation of "background" signal and hence suppression of significant features at the given position.
Version 1.3 beta:
- Progress bars now appear for long-running tasks such as loading peptide file and generating logos and logo trees
- Notable bug fixes:
- Selected profiles now correctly closed with the "Close Selected" button.
- Regenerating a logo tree after closing some profiles now correctly excludes the closed profiles.
Version 1.2 beta:
- Logo trees can be generated for all types of domains - not only terminal binding
- New advanced options including amino acid coloring styles, amino acid grouping, and logo tree leaf ordering
Version 1.1 beta:
New Logo Tree feature - basic functionality for PDZ or other terminal binding motifs not requiring profile alignment
Installing and Running
Requirements: Java Runtime Environment (JRE) 1.5 or later is required to run LOLA. All other dependencies are included in the download.
- Extract the TAR file. This will create a directory named "lola".
- On Linux, open the command shell and run "lola.sh" from the "lola" directory.
- On Windows, double click "lola.bat".
- On Mac, double click lola-1.0-beta.jar
LOLA accepts one or more peptide file as shown in the example below. A peptide file describes a protein containing a specific domain, and provides known peptide ligands of this domain obtained by an experimental technique. You can open a single peptide file, or multiple peptide files grouped into a project file.
LOLA also accepts one or more pwm file as shown in the example below. A pwm file describes a position weight matrix (rows are amino acids, columns are ligand positions) where each cell in the matrix represents the frequency of observing a specific amino acid at a given ligand position. You can open a single pwm file, or multiple pwm files grouped into a project file.
Here's a view of LOLA after opening a PDZ domain project file, and after generating a logo tree:
Changing memory allocations on Windows, Mac, and Linux machines
There are a number of ways to change LOLA's memory allocation, depending on your preferred method of opening the application.
Option A: Command line startup (note: this does not permanently change LOLA's default 512M setting)
If you are opening LOLA from the command-line using the command:
- java –Xmx512M –jar lola-VERSION.jar
then you can increase the value of –Xmx to the desired amount of memory. For example:
- java –Xmx800M –jar lola-VERSION.jar
Option B: Using lola.bat (Windows systems)
- Open the file lola.bat in a text editor (eg. right-click and select Open With Notepad).
- Increase the value of the –Xmx tag (found in the last line of the file), as per Option A. Do not modify other parts of the file.
- Save and close the file.
- Open LOLA by double-clicking on lola.bat.
Option C: Using lola.sh (UNIX, Linux, and Mac OS X systems)
Open the file lola.sh in a text editor (eg. right-click and select Open With TextEdit).
- Increase the value of the –Xmx tag (found in the last line of the file), as per Option A. Do not modify other parts of the file.
- Save and close the file.
- Open LOLA by running lola.sh from the command-line.
References
LOLA has been referenced in the following publications:
Tonikian, R.
et al.(2008) A Specificity Map for the PDZ Domain Family.
PLOS Biology, 6, 9: 2043-2059.
http://biology.plosjournals.org/perlserv/?request=get-document&doi=10.1371/journal.pbio.0060239
Future Developments
- Enable logo and logo tree parameters to be saved in a project file for later use
- Support customized amino acid grouping schemes
- Allow colours to be selected for individual amino-acids
- Add support for nucleic acids
- Additional visualization options (e.g. font, axis labels)
Contact
If you have any questions or feedback, please email Moyez Dharsee at mdharsee@infochromics.com .